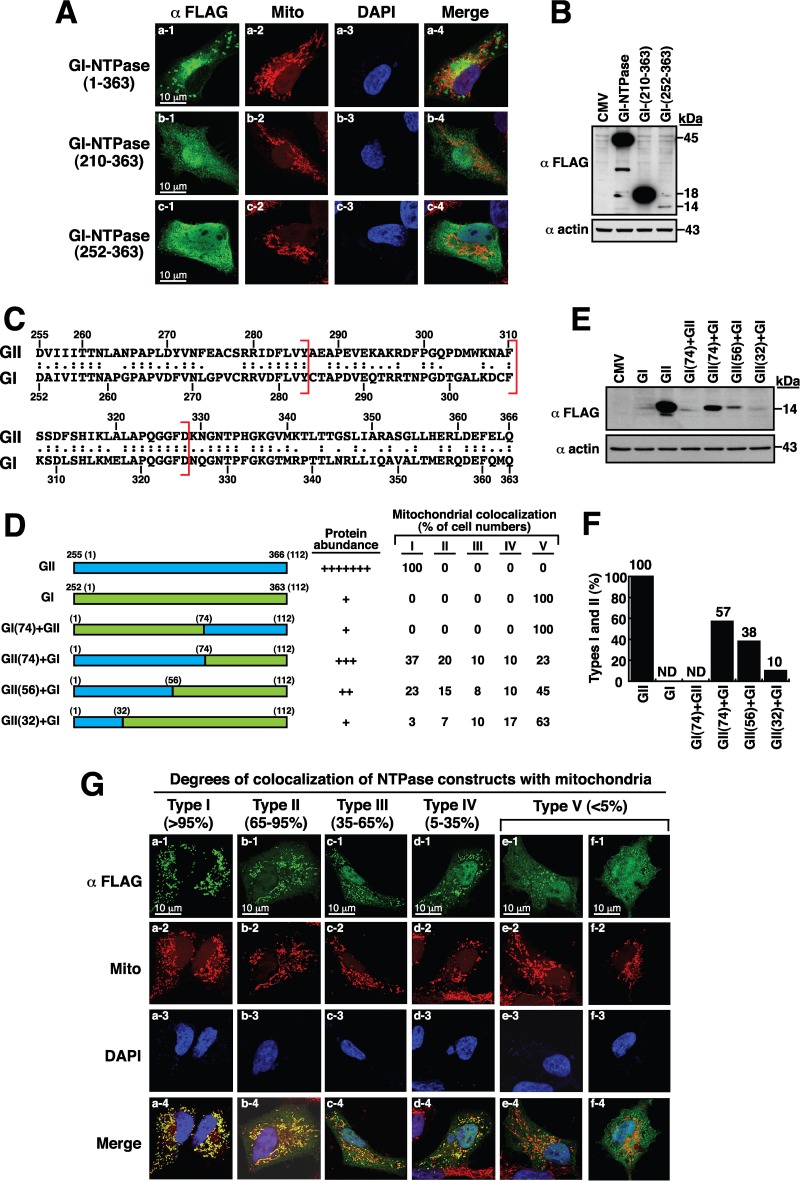
FIG 5.
Full-length and N-terminal-deleted NTPase constructs from a GI norovirus strain do not show significant colocalization with mitochondria. (A) Immunofluorescence analysis of the subcellular localization of the full-length and N-terminal-deleted GI-NTPase constructs in A7 cells. As noted, GI-NTPase(1–363), GI-NTPase(210–363), and GI-NTPase(252–363) did not show substantial colocalization with mitochondria. Scale bars, 10 μm. (B) Western blot analysis of the full length and deletion constructs of GI-NTPase expressed in 293T cells. (C) Sequence alignment of amino acid residues between GII-NTPase from aa 255 to 366 and GI-NTPase from aa 252 to 363. (D) Diagram of domain-swapped NTPase constructs and summary of their protein abundance and mitochondrion-targeting ability. (E) The degrees of protein abundance for each NTPase chimeric construct, determined by Western blotting, are indicated by a plus sign. As noted, several domain-swapped NTPase constructs produced diversified phenotypes with various degrees of mitochondrial colocalization in transfected cells. (G) The heterogeneous cell populations were divided into five types (types I to V) according to the degrees of NTPase chimeric constructs colocalized with the mitochondria. Cells with the type I, II, III, IV, and V patterns were defined as >95%, 65 to 95%, 35 to 65%, 5 to 35%, and <5% portions, respectively, of NTPase constructs colocalized with the mitochondria. Scale bars, 10 μm. (F) The percentages of the type I and type II patterns in transfected cells for the indicated NTPase constructs were evaluated by counting at least 300 FLAG-positive cells (n > 300).