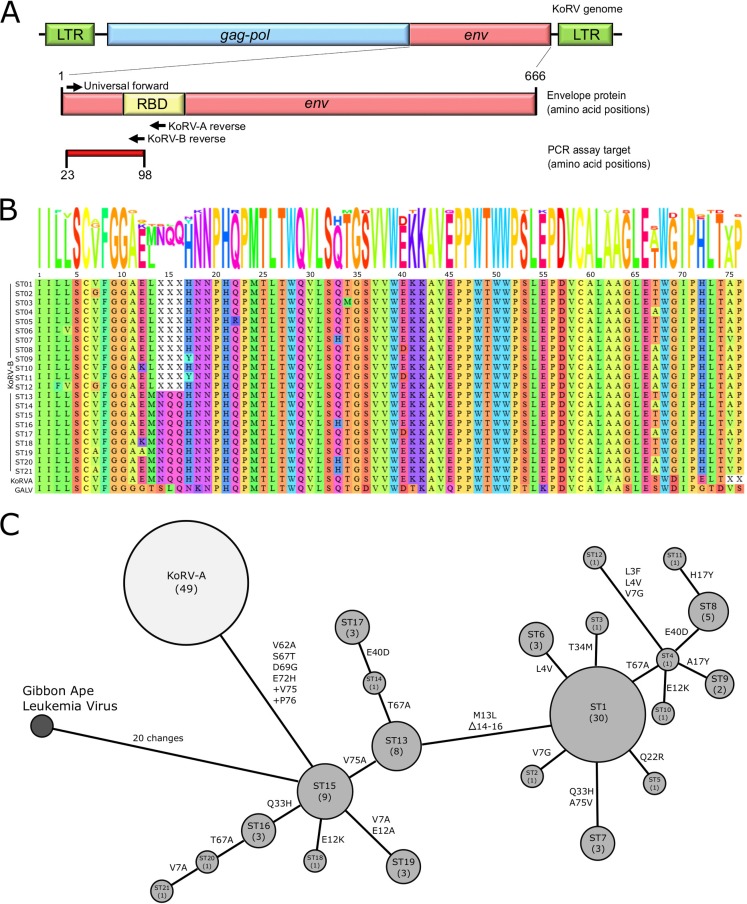
FIG 1.
KoRV sequence variation in the MBR (Moreton Bay Rail) population. (A) Schematic of KoRV genome highlighting the targeted region of the envelope (env) gene for analysis. LTR, long terminal repeat; RBD, receptor binding domain. (B) Alignment of unique KoRV envelope fragments detected. The fragment represents amino acids 23 to 98 in the full-length sequence. One KoRV-A sequence was detected, while 21 unique KoRV-B sequence types (ST) were detected. Gibbon ape leukemia virus (GALV) is included as an outlier. Gaps are represented by “X,” and the alignment sequence logo is shown above. (C) Minimum spanning tree representing the KoRV envelope protein sequence. Medium gray nodes represent KoRV-B, with the ST indicated and the number of individual koalas with that ST stated in parentheses. Amino acid changes between node sequences are indicated on vertices.