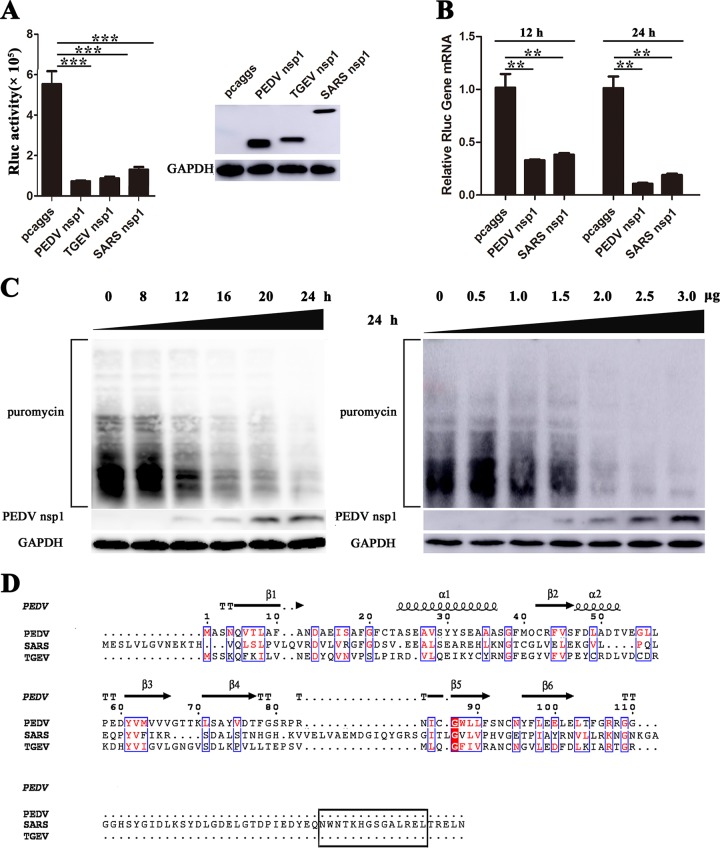
FIG 4.
CoVs nsp1 inhibit host protein synthesis. (A) PK-15 cells were cotransfected with pRL-TK carrying the Rluc reporter gene downstream of the TK promoter and one of the plasmids pcaggs, pcaggs-PEDV-nsp1-HA (PEDV nsp1), pcaggs-TGEV-nsp1-HA (TGEV nsp1), or pcaggs-SARS-CoV-nsp1-HA (SARS nsp1), which express PEDV nsp1, TGEV nsp1 and SARS-CoV nsp1, respectively. All the expressed proteins had N-terminal HA tags. At 24 h posttransfection, cell lysates were prepared and subjected to Rluc assays. Error bars show the standard deviations (SDs) of the results from three independent experiments. Cell extracts were also subjected to Western blot analysis using an anti-HA antibody (top) or an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (bottom). *, P < 0.05 (considered statistically significant); **, P < 0.01 (considered highly significant); ***, P < 0.001 (considered extremely significant). (B) HEK-293T cells were transfected with the wild-type plasmid. At 12 or 24 h posttransfection, the cells were lysed and subjected to real-time quantitative PCR analysis. Asterisks indicate statistical significance calculated by the Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) HEK-293T cells were transfected with the pcaggs-PEDV-nsp1-HA (PEDV nsp1) plasmid. Left, the cells were pulsed with 3 μM puromycin for 1 h at 0, 8, 12, 16, 20, and 24 h posttransfection and then subjected to Western blot analysis. Right, different doses of the wild-type plasmid (0 to 3.0 μg) were transfected into HEK-293T cells. After 24 h posttransfection, the cells were pulsed with 3 μM puromycin for 1 h at 37°C and then subjected to Western blot analysis. (D) The amino acid sequences of PEDV nsp1, SARS-CoV nsp1, and TGEV nsp1 were compared, and the functional region (aa 160 to 173) of SARS-CoV nsp1 is indicated with a black box.