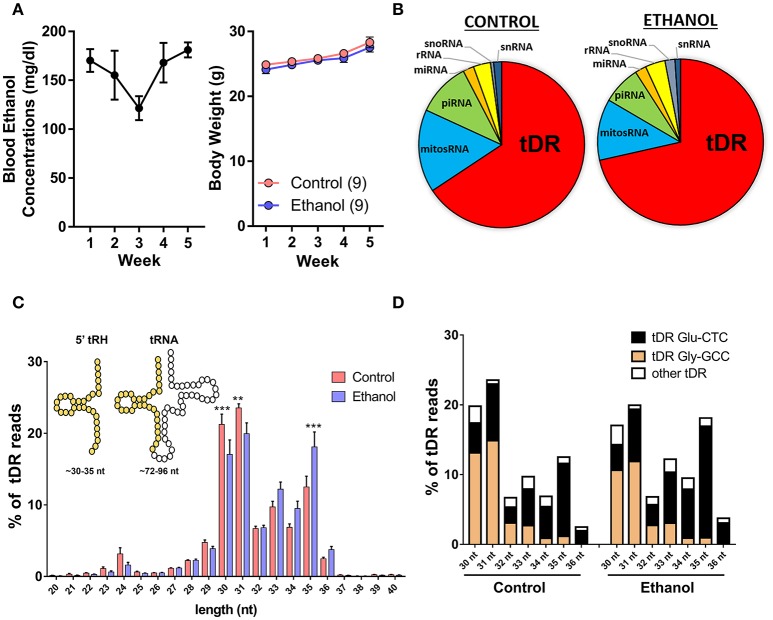
Figure 1.
Chronic ethanol shifts the tDR profile of sperm small noncoding RNA. (A) Chronic intermittent ethanol vapor exposure (left panel) induced an average blood ethanol concentration of 159.2 ± 9.2 mg/dl [mean (μ) ± standard error of the mean (SEM)] over its 5 week duration. There was no effect of chronic ethanol on body weight (right panel) compared to the control group (p > 0.05). (B) Pie charts displaying the percentage of each small RNA class represented in sperm from control and ethanol treatment groups. (C) Most tDR are 30–35 nt 5′-derived tRNA halves (5′-tRH) (see insert) and chronic ethanol significantly altered the percentage of 30, 31, and 35 nt tDR reads. (D) Most 30–36 nt tDR reads map to Glu-CTC and Gly-GCC relative to all other tDR species. Data in bar graphs presented as μ ± SEM. N = 9/treatment in all panels. **p < 0.01. ***p < 0.001.