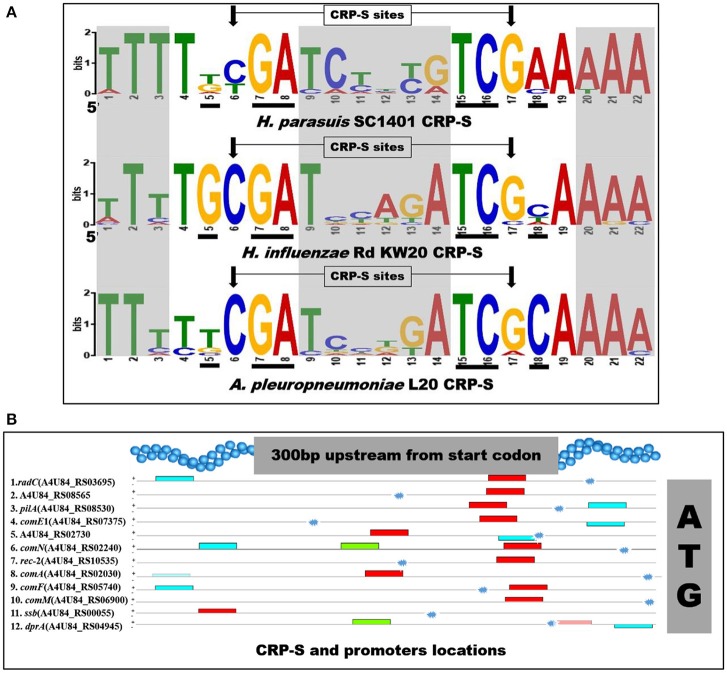
Figure 2.
Sequences logos of CRP-S in H. parasuis SC1401 and H. influenzae Rd KW20 and locations in upstream of 11 competence genes-homologs in SC1401. (A) CRP-S logos of 12 H. parasuis sites (top, with an E-value of 5.3e-035) and 13 H. influenzae sites (middle), as well as 15 A. pleuropneumoniae sites (bottom). The motifs were found and logos generated with MEME (http://meme-suite.org/tools/meme) (Bailey and Elkan, 1994). Bases important for cAMP-CRP-DNA binding reported by Prof. Redfield are shown in black underlines, and the characteristic positions of CRP-S sites (C6/C17) are shown in arrows. Noncore sites are shaded. Logo of CRP-S of H. influenzae was reproduced with Prof. Redfield's permission. (B) CRP-S locations in 12 H. parasuis competence genes. Red boxes indicate 22 bp consensus sequences with high similarity with CRP-S shown in Table 3 and Figure 2 (top); the green and blue ones indicate similar motifs. Note, the CRP-S motif predicted in promoter region of dprA was partly homologous, thus indicated by a semitransparent red box. Promoter of each gene is shown with explosion icon, which was predicted with BPROM (http://linux1.softberry.com/berry.phtml).