Fig. 1.
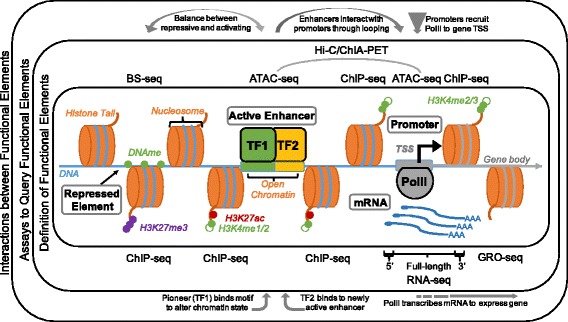
Common functional elements. Inner box: definitions of various functional elements (left to right): repressed element with methylated DNA (DNAme) and repressive modification of the histone tail of a nearby nucleosome (H3K27me3); active enhancer in an open chromatin region bound by two transcription factors (TF1, TF2) and marked by H3K4me1 or H3K4me2 (enhancer) and H3K27ac (activity); promoter bound by RNA Polymerase II (PolII) in an open chromatin region around the transcription start site (TSS, black arrow) of a gene body and marked by H3K4me2 or H3K4me3; and mRNA molecules transcribed from the gene with 5′ caps and 3′ poly (A) tails. Middle box: list of high-throughput sequencing assays used to annotate these functional elements, including bisulfite sequencing (BS-seq) for DNA methylation, Assay for Transposase Accessible Chromatin (ATAC-seq) for open chromatin, high-throughput chromosome conformation capture (Hi-C) or Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET) for interactions between regions, chromatin immunoprecipitation (ChIP-seq) for histone modifications, RNA-sequencing (RNA-seq) for mature mRNA (either 5′ biased, full length, or 3′ biased), and Global Run-On (GRO-seq) for nascent mRNA. Outer box: interactions between functional elements
