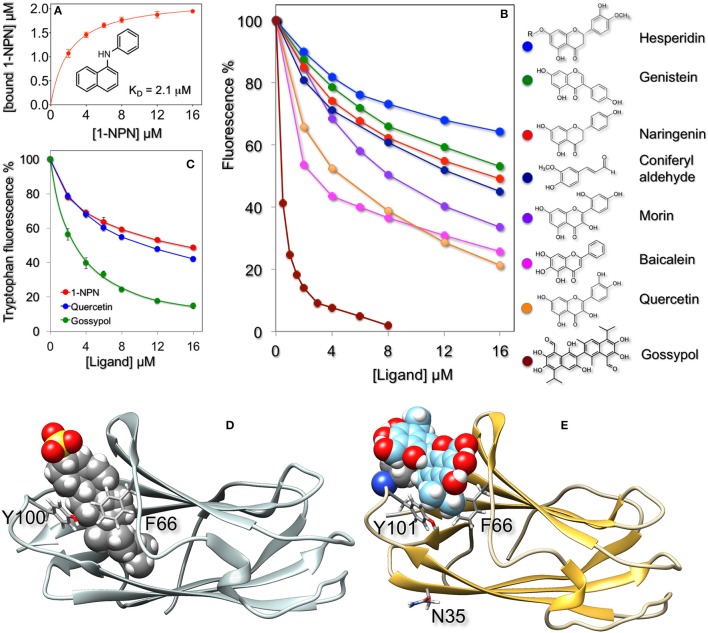
Figure 2.
Ligand-binding properties of recombinant HarmNPC2-1. (A) The purified protein binds the fluorescent probe 1-NPN with a dissociation constant of 2.1 μM (SEM: 0.14). (B) In competitive binding experiments a series of flavonoids exhibited good affinity for HarmNPC2-1. Among natural volatile compounds only coniferyl aldehyde showed moderate binding. Similar affinity was measured with synthetic derivatives (α-methyl, α-methoxy, and α-pentyl) of cinnamaldehyde. The sex pheromone components of H. armigera (Z)-11-hexadecenal, (Z)-9-hexadecenal, (Z)-11-hexadecenol and (Z)-11-hexadecenyl acetate, as well as the following chemicals were ineffective in displacing the fluorescent probe from the complex at concentrations up to 16 μM: benzaldehyde, 4-methyl benzaldehyde, 2-hydroxy benzaldehyde, 4-methoxy benzaldehyde, vanillin, cinnamaldehyde, eugenol, piperonyl alcohol, 2-phenylacetaldehyde, safranal, carvone, citronellal, citronellic acid methyl ester, methyl cinnamate, methyl jasmonate, β-ionone, capsaicin, farnesol, indole, β-estradiol, 1-octen-3-ol, octanal, 2,4-octadienal, octanoic acid, decanal, decanoic acid, Z3-hexen-1-ol, 2-hexanol, 3-hexanol, 1-heptanol. (C) Quenching of intrinsic tryptophan fluorescence by 1-NPN, quercetin, and gossypol. (D) Docking of cholesterol sulfate (shown in fill mode) to bovine NPC2 (PDB: 2HKA) and of gossypol to a model of HarmNPC2-1 (E). In both cases the ligand binds in the same region of the protein and is flanked by the same residues, Phe66 and Tyr100/Tyr101, shown in stick mode. In the model of HarmNPC2-1 the predicted glycosilation site at Asn3 is also shown in stick mode. The model of HarmNPC2-1 was built using the on-line programme SwissModel (Peitsch, 1995; Arnold et al., 2006; Kiefer et al., 2009) and the structure of the human NPC2 (PDB: 5KWY, Li et al., 2016) as template.