Figure 7.
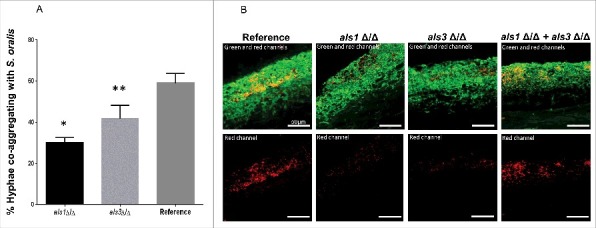
Als1 promotes co-aggregation interactions between C. albicans and S. oralis. (A) Co-aggregation assays between S. oralis 34 and C. albicans reference strain or strains lacking either ALS1 (als1Δ/Δ) or ALS3 (als3Δ/Δ) were performed as described in methods. Results represent percentage of hyphae co-aggregating with S. oralis in each microscopic field, at 40X magnification. Means ± SD are shown from 3 independent experiments, with 8 microscopic fields analyzed per condition in each experiment. Deletion of ALS1 or ALS3 significantly decreased the number of hyphae co-aggregating with S. oralis compared with the reference strain. *p<0.0001, **p<0.05 for a comparison with the reference strain. (B) Mucosal biofilms of C. albicans with S. oralis growing for 16 h on the surface of oral mucosal organotypic constructs. Tissues were inoculated with S. oralis 34 together with a C. albicans reference strain, als1Δ/Δ mutant, als3Δ/Δ mutant or a 1:1 mixture of the 2 mutants. Fluorescence images (top panel) show biofilms labeled with a FITC-conjugated anti-Candida antibody (green) and an Alexa Fluor 568-labeled streptococcal FISH probe for S. oralis (red). The red channel is individually shown in the lower panel to better visualize the bacterial biomass. Mixing the 2 mutants restored the S. oralis biofilm on mucosal surfaces, supporting a complementary functional role of Als1 and Als3 adhesins in cross-kingdom co-aggregation interactions. Bars: 50 µm.
