Figure 8.
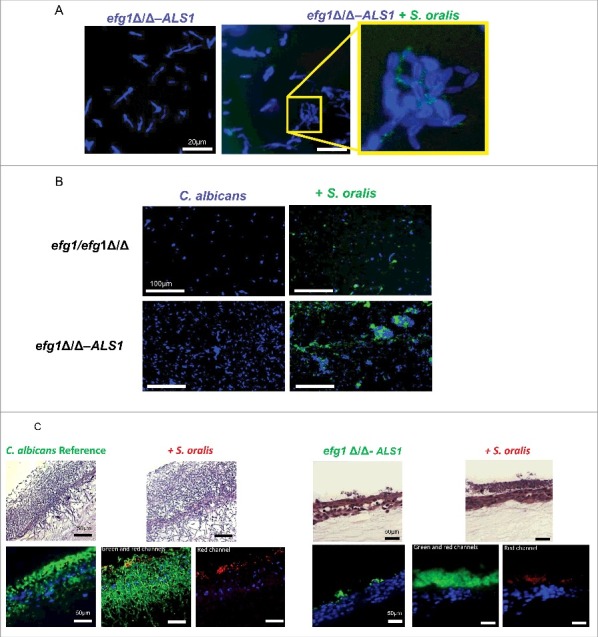
Overexpression of the ALS1 gene in the efg1Δ/Δ background partially rescues the cross-kingdom mucosal biofilm phenotype (A-B) Co-culture of a C. albicans ALS1-overexpressing strain in the efg1Δ/Δ background (efg1Δ/Δ-ALS1) with a teal protein expressing S. oralis 34 strain for 45 minutes. (A) or 24 hours (B) on Permanox® plastic chamber slides, in RPMI 10%FBS, 10%BHI media. Candida cells were stained with Calcofluor White (blue) and filamentation pattern and co-aggregation interactions were observed under a fluorescence microscope. A representative of 2 experiments is shown, with conditions set up in duplicate. Fungal-bacterial cell co-aggregation interactions were observed both in early (Fig. 8A) and late (Fig. 8B) stages of mixed biofilm development with the ALS1 overexpressing strain. Bars: 20 μm (A); 100 μm (B). (C) Sixteen-hour mucosal biofilms of C. albicans (left panels) or C. albicans with S. oralis (right panels). Biofilms of the reference strain and the efg1Δ/Δ-ALS1 overexpressing strain, with or without S. oralis 34, were grown on the surface of organotypic oral mucosal surfaces. H&E staining (top panels) and fluorescence images of biofilms labeled with a FITC-conjugated anti-Candida antibody (green), an Alexa Fluor 568-labeled streptococcal FISH probe for S. oralis (red), and counterstained with the nucleic acid stain Hoechst 33258 (blue) to visualize mucosal cells, are shown. Green and red channels are individually shown in the efg1Δ/Δ-ALS1 plus S. oralis panels to better visualize the fungal and bacterial signals. A representative of 2 experiments is shown, with conditions set up in duplicate. Overexpression of ALS1 in the efg1Δ/Δ strain background restored S. oralis biofilm to levels similar to the reference and efg1 revertant strains (see Fig. 5A for comparison to the revertant strain). Bars: 50 µm.
