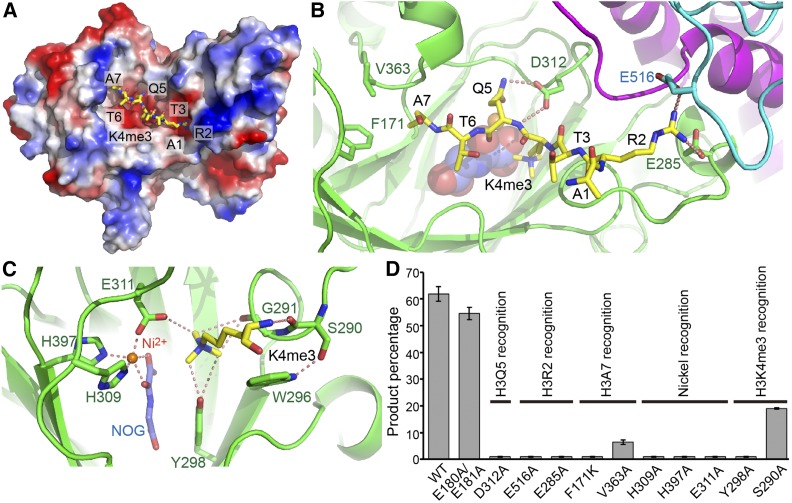
Figure 3.
Interactions between JMJ14 and H3K4me3 Peptide.
(A) An electrostatics surface view of JMJ14CD with the H3K4me3 peptide shown in stick representation. The peptide docks in a negatively charged surface cleft of JMJ14.
(B) The specific recognition of H3R2, H3Q5, and H3A7 by JMJ14. The H3R2 side chain inserts into a binding pocket at the junction formed by jumonji, helical, and C5HC2 domains and forms hydrogen bonding and salt bridge interactions with Glu-285 and Glu-516. The hydrogen bonds are highlighted by dashed red lines. H3Q5 forms two hydrogen bonds with Asp-312. The side chain methyl group of H3A7 inserts into a shallow hydrophobic pocket formed by Phe-171 and Val-363.
(C) The specific recognition of H3K4me3 and the active site conformation. The H3K4me3 main chain forms a hydrogen bond with Ser-290. The methyl groups of the trimethyllysine are specifically recognized by a CH-O hydrogen bonding network. The nickel ion is coordinated by His-309, Glu-311, His-397, and the cofactor analog NOG.
(D) The quantified in vitro demethylation assay of JMJ14CD and its mutants. The percentages of the product peptide are shown as means ± sd (n = 3). The functional annotations of the mutation sites are indicated above the transverse lines. Mutations of important residues involved in substrate binding are shown to significantly impair activity.