The essential rRNA processing factor APUM24 functions in removal of internal transcribed spacer 2, and reduction of APUM24 causes sugar-dependent rRNA processing defects and nucleolar stress.
Abstract
Ribosome biogenesis is one of the most energy-consuming events in the cell and must therefore be coordinated with changes in cellular energy status. Here, we show that the sugar-inducible gene ARABIDOPSIS PUMILIO PROTEIN24 (APUM24) encodes a Pumilio homology domain-containing protein involved in pre-rRNA processing in Arabidopsis thaliana. Null mutation of APUM24 resulted in aborted embryos due to abnormal gametogenesis and embryogenesis, whereas reduced expression of APUM24 caused several phenotypes characteristic of ribosome biogenesis or function-related mutants. APUM24 interacted with other pre-rRNA processing factors and a putative endonuclease for the removal of the internal transcribed spacer 2 (ITS2) of pre-rRNA in the nucleolus. The APUM24-containing complex also interacted with ITS2, and reduced APUM24 expression caused the overaccumulation of processing intermediates containing ITS2. Thus, APUM24 likely functions as an ITS2 removal-associated factor. Most importantly, the apum24 knockdown mutant was hypersensitive to highly concentrated sugar, and the mutant showed sugar-dependent overaccumulation of processing intermediates and nucleolar stress (changes in nucleolar size). Furthermore, reduced APUM24 expression diminished sugar-induced promotion of leaf and root growth. Hence, a breakdown in the coordinated expression of ribosome biogenesis-related genes with energy status may induce nucleolar stress and disturb proper sugar responses in Arabidopsis.
INTRODUCTION
The eukaryotic ribosome is composed of two subunits, namely, the 60S large subunit and the 40S small subunit. The large subunit comprises 25S, 5.8S, and 5S rRNAs and ∼47 ribosomal proteins (RPs), whereas the small subunit comprises 18S rRNA and ∼33 RPs (Henras et al., 2015; Weis et al., 2015a). Ribosome biogenesis begins with the biosynthesis of 45S pre-rRNA by RNA polymerase I in the nucleolus. This long transcript is processed into 18S, 5.8S, and 25S rRNA via the truncation of 5′ and 3′ external transcribed spacers (5′ETS and 3′ETS) and the removal of internal transcribed spacer 1 and 2 (ITS1 and ITS2). The 45S pre-rRNA functions as a scaffold for the assembly of 5S rRNA transcribed by RNA polymerase III in the nucleoplasm, RPs, numerous ribosome biogenesis factors (RBFs), and small nucleolar RNAs in the nucleolus to form the 90S preribosomal particle. During the maturation step, the 45S pre-rRNA in the 90S preribosomal particle is processed and the particle is divided into 40S and 60S preribosome subunits. These subunits are exported into the cytoplasm and reassembled, thereby becoming functional in the cytosol. Since rRNA synthesis is the rate-limiting step of ribosome biogenesis, its modulation plays a central role in controlling ribosome biogenesis (Kressler et al., 2010; Tschochner and Hurt, 2003).
Since ribosome biogenesis is one of the most energy-consuming processes in the cell, it must be controlled in coordination with changes in intracellular energy status (Lempiäinen and Shore, 2009). In fact, sugar deprivation treatments reduce rRNA synthesis in both mammalian and plant cells (Murayama et al., 2008; Ishida et al., 2016), while exogenous sugar application increases the expression levels of a number of genes associated with ribosome biogenesis in Arabidopsis thaliana (Kojima et al., 2007), as well as yeast and mammalian cells (Powers and Walter, 1999; Iadevaia et al., 2014). Since ribosome biogenesis consists of multiple processes, including pre-rRNA synthesis, RP and RBF production, the assembly of these components, and pre-rRNA processing, these processes must be coordinately modulated in response to the intracellular sugar status. Although the mechanism connecting sugar status (energy status) with ribosome biogenesis has not yet been fully elucidated, several factors involved in this mechanism have been identified. For instance, a target of rapamycin (TOR) kinase and its downstream factor RPS6 kinase (S6K) play critical roles in energy signaling in yeast and mammalian cells (Loewith and Hall, 2011; Jewell and Guan, 2013). The TOR-S6K signaling pathway was also recently shown to accelerate the transcription of not only rRNA but also many RP genes in plant cells (Xiong et al., 2013; Xiong and Sheen, 2014).
The ribosome is required for translation, making it absolutely essential for maintaining the vitality of the cell. Therefore, defects in ribosome biogenesis or function exert pleiotropic effects on all cellular functions (Byrne, 2009). However, some distinct mutations in human RP and RBF genes lead to different but specific diseases known as ribosomopathies through an almost completely unknown mechanism (Chakraborty et al., 2011; Armistead and Triggs-Raine, 2014). Ribosome biogenesis is also strongly associated with tumor suppression through the nucleolar stress response (also known as the ribosome stress response) in mammals (Holmberg Olausson et al., 2012; Zhou et al., 2015). Nucleolar stress, i.e., the disruption of the nucleolar structure caused by a failure in ribosome biogenesis, leads to cell cycle arrest, senescence, and apoptosis (Boulon et al., 2010; Nicolas et al., 2016). The RP-mouse double minute2 (MDM2)-p53 pathway, including the p53-targeted ubiquitin ligase MDM2 and the tumor suppressor protein p53, plays a central role in controlling the nucleolar stress response in mammals (Zhou et al., 2015). However, other pathways might also be involved in the nucleolar stress response. In fact, the nucleolar stress response also occurs in yeast cells lacking homologs of mammalian p53 and MDM2 proteins (James et al., 2014). The effects of defects in ribosome biogenesis or function in plants have been investigated by phenotypic analyses of Arabidopsis mutants with reduced expression of RP or RBF. Such defects frequently cause abnormal gametogenesis, embryogenesis, seed germination, root elongation, and leaf morphology (Byrne, 2009; Horiguchi et al., 2012; Weis et al., 2015a). Hence, the regulation of ribosome biogenesis and function is likely associated with plant-specific physiological processes throughout the entire plant life cycle.
Pum and FBF (PUF) proteins contain a Pumilio homology domain (PUM-HD), which generally functions as an RNA binding domain and is universally conserved in eukaryotic organisms (Spassov and Jurecic, 2003). The canonical PUM-HD consists of eight PUF repeats in tandem, each of which recognizes a single nucleotide in its target RNA (Spassov and Jurecic, 2003). Typical yeast and Caenorhabditis elegans PUF proteins bind to specific recognition sequences in the 3′-untranslated regions of their mRNA targets to modulate their translational efficiency or stability (Wickens et al., 2002). However, PUF proteins have not yet been characterized in plants, except for a few Arabidopsis PUF proteins. The Arabidopsis genome encodes 25 or 26 PUF proteins that form a protein family designated the Arabidopsis Pumilio protein (APUM) family (Francischini and Quaggio, 2009; Tam et al., 2010). Recently, APUM5 was found to regulate Cucumber mosaic virus infection and abiotic stress responses through direct binding to Cucumber mosaic virus RNA and the 3′-untranslated regions of mRNAs from abiotic stress-responsive genes (Huh et al., 2013; Huh and Paek, 2014). APUM9 controls seed dormancy, functioning as a downstream factor of the dormancy regulatory genes REDUCED DORMANCY5 and DELAY OF GERMINATION6 (Xiang et al., 2014, 2016). On the other hand, it was recently suggested that APUM23 functions in rRNA processing by directly binding to a 10-nucleotide sequence in 18S rRNA (Zhang and Muench, 2015).
APUM24 and its rice homolog were initially identified as the nuclear glucose-inducible gene-encoded Armadillo/Pumilio domain-containing proteins AtNuGAP1 and OsNuGAP1 (Aki and Yanagisawa, 2009), and APUM24 was subsequently identified as a member of the APUM family (Francischini and Quaggio, 2009). APUM24 is localized to the nucleolus (Tam et al., 2010) and has an amino acid sequence that is quite different from those of other APUM proteins (Francischini and Quaggio, 2009). Furthermore, unlike other APUM proteins, APUM24 contains an imperfect PUM-HD sequence. Thus, APUM24 might play a unique role in Arabidopsis. In this study, to uncover the molecular function and physiological role of APUM24, we performed a variety of phenotypic analyses with Arabidopsis plants harboring a null or knockdown mutation of APUM24 as well as proteome analysis of the APUM24-containing protein complex. Our results suggest that APUM24 is an essential gene encoding a pre-rRNA processing-associated factor necessary for removal of ITS2. Moreover, reduced expression of the sugar-inducible gene APUM24 induced nucleolar stress in a sugar-dependent manner, led to hypersensitivity to high concentrations of sugar, and largely diminished sugar-induced promotion of leaf and root growth. Despite the many studies on nucleolar stress in yeast and mammals, nucleolar stress in plants has rarely been investigated. Hence, our findings uncover links among ribosome biogenesis, nucleolar stress, and sugar responses in plants and highlight the physiological importance of nucleolar stress in plant cells.
RESULTS
Null and Knockdown Mutations of APUM24 Cause Abortion and Delay in Embryogenesis
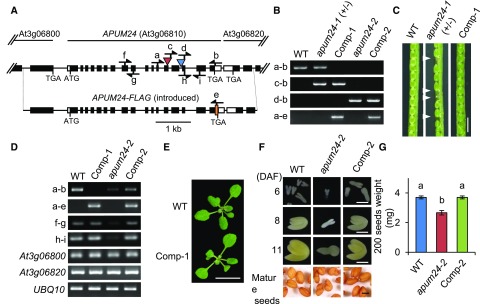
To reveal the physiological roles of APUM24, we examined the phenotypes of two Arabidopsis T-DNA insertion lines. Since a T-DNA insertion is located within the 13th exon and 14th intron of APUM24 (At3g06810) in lines GABI_461E08 and SALK_033623, respectively, these mutant alleles were designated apum24-1 and apum24-2 (Figure 1A). As described below, apum24-1 and apum24-2 are null and knockdown mutant alleles of APUM24, respectively.
Figure 1.
Null and Knockdown Mutations of APUM24 Cause Embryonic Lethality and Delayed Embryogenesis.
(A) Schematic representation of the native APUM24 allele and the APUM24 allele introduced by transformation (APUM24-FLAG). White and black boxes indicate untranslated and coding regions in exons for APUM24, respectively. Red and blue triangles indicate the positions of T-DNA insertions in the apum24-1 and apum24-2 mutant alleles, respectively. “ATG” and “TGA” indicate the translational initiation and stop codon, respectively. The orange box is a region encoding three copies of FLAG tag. Arrows indicate the positions of primers used for genotyping or RT-PCR.
(B) to (G) Wild-type plants, a heterozygote for the apum24-1 allele [apum24-1(+/−)], a homozygote for the apum24-2 allele (apum24-2), and transgenic lines homozygous for the apum24-1 or apum24-2 alleles but harboring an introduced wild-type APUM24 allele (Comp-1 and Comp-2) were used.
(B) PCR-based genotyping. Primers indicated in (A) were used for PCR amplification of the native APUM24 (a-b), apum24-1 (c-b), apum24-2 (d-b), and introduced APUM24 alleles (a-e), respectively.
(C) Photographs of seeds in siliques. Arrowheads indicate aborted embryos. Bar = 1 mm.
(D) RT-PCR analysis of APUM24 transcripts from the native and introduced APUM24 loci. Primers indicated in (A) were used for PCR-based specific detection of transcripts from the native (a-b) and introduced APUM24 loci (a-e). Transcripts containing the region upstream (f-g) or downstream (h-i) of the T-DNA insertion sites, and transcripts from neighboring genes of APUM24 (At3g06800 and At3g06820) were also analyzed. UBQ10 was used as an internal control.
(E) Photographs of the seedlings grown on 1/2 MS plates for 2 weeks. Bar = 1 cm.
(F) Photographs of seeds and isolated embryos. DAF, days after flowering. Bar = 1 mm.
(G) The 200-seed weight values. Error bars indicate sd (n = 3). Statistical significance was determined by ANOVA, followed by a Tukey-Kramer test. Means that are significantly different from each other (P < 0.05) are indicated by different letters.
The apum24-1 heterozygotes (Figure 1B) grew normally during the vegetative phase; however, they developed aborted seeds (Figure 1C), suggesting that apum24-1 is defective in gametogenesis and/or embryogenesis. We therefore backcrossed the heterozygote to wild-type plants three times and investigated the segregation ratio of apum24-1 in the progeny of the backcrossed line after self-pollination. After self-pollination, we failed to obtain any plants homozygous for apum24-1, and the ratio of wild-type to heterozygous plants in the progeny population was very different from 1:2 (Table 1; χ2 test, P = 0.004). Thus, apum24-1 is likely impaired in both gametogenesis and embryogenesis.
Table 1. Segregation of apum24-1 (+/−) Self-Progeny and Reciprocal Crosses between apum24-1 (+/−) and the Wild Type (+/+).
| Parental Genotypes (Female × Male) | Genotypes of F1 Plants (n) | Total (n) | Transmission Efficiency (%)a | χ2 Test (P) | |||
|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||||
| +/− × +/− | 115 | 163 | 0 | 278 | 141.7 | 0.004b | (vs. 1:2) |
| +/+ × +/− | 78 | 66 | 0 | 144 | 84.6 | 0.317 | (vs. 1:1) |
| +/− × +/+ | 88 | 55 | 0 | 143 | 62.5 | 0.005b | (vs. 1:1) |
Transmission efficiency (%) = (mutant/wild type) × 100.
Significant difference (P < 0.05).
To clarify the relationship between the apum24-1 allele and defects in gametogenesis and embryogenesis, we generated transgenic plants homozygous for apum24-1 by introducing a genomic DNA fragment containing the APUM24 locus into an apum24-1 heterozygote (Figure 1B). The genomic DNA contained a 1.2-kb region upstream of the transcription start site, the entire transcribed region, and a 1-kb region downstream of the polyadenylation site, as well as a FLAG-tag-encoding sequence in front of the stop codon of the APUM24 coding region (Figure 1A). We established the transgenic line (line Comp-1) by genotyping (Figure 1B) and examined transcripts from the introduced and native APUM24 genes in the T3 progenies using RT-PCR analysis with appropriate primers (Figures 1A and 1D). Transcripts from the introduced APUM24 gene (Figure 1D, panel a-e) but not from the native APUM24 gene (panel a-b) were detected in this line. The levels of expression of the endogenous APUM24 gene in the wild type were comparable to those of the introduced APUM24 gene in the transgenic line (Figure 1D, panels f-g and h-i). Because the introduced DNA included parts of neighboring genes from upstream and downstream of APUM24 (At3g06800 and At3g06820), we also confirmed that the expression levels of these genes were not affected in the Comp-1 line (Figure 1D). All embryos developed normally into seeds in the siliques of this transgenic line (Figure 1C), and the growth of the transgenic line was similar to that of the wild type throughout the vegetative phase (Figure 1E). Therefore, apum24-1 is a null allele that causes lethality in the generative phase of growth. The ratio of wild-type to heterozygous plants was 1:1 (χ2 test, P = 0.317) in a progeny population produced by crossing wild-type plants with an apum24-1 heterozygote as the paternal parent (Table 1). However, when the apum24-1 heterozygote was used as the maternal parent, the segregation ratio was not 1:1 (χ2 test, P = 0.005), and the transmission efficiency was 62.5% (Table 1). These results indicate that the apum24-1 allele negatively affects female gametogenesis as well as embryogenesis.
On the other hand, homozygotes for the apum24-2 allele (Figure 1B) were viable. Therefore, the homozygote is referred to as the apum24-2 mutant hereafter. In apum24-2 mutant seedlings, APUM24 transcripts were detectable but at levels much lower than those in wild-type seedlings, indicating that the apum24-2 mutant is a knockdown mutant (Figure 1D, panel a-b; Supplemental Figure 1A). RT-PCR analysis using two sets of primers for amplification of the regions upstream and downstream of the inserted T-DNA demonstrated that the T-DNA insertion did not cause the accumulation of truncated APUM24 transcripts (Figure 1D, panels f-g and h-i). Consistent with the severe effect of the apum24-1 null allele on embryogenesis, the apum24-2 mutant developed wrinkled seeds (Figure 1F), and the seed weight of the apum24-2 mutant was lighter than that of the wild type (Figure 1G). To investigate this phenotype in more detail, we observed embryos at various stages of embryogenesis and found that embryogenesis was delayed in the apum24-2 mutant (Figure 1F). By introducing the wild-type APUM24 locus into the apum24-2 mutant, we generated a complementation line (line Comp-2) (Figures 1B and 1D). Because the introduction of the wild-type APUM24 locus rescued both the delayed embryogenesis and abnormal seed shape of the mutant (Figures 1F and 1G), we reasoned that the wrinkled seeds of the apum24-2 mutant were probably caused by delayed embryogenesis. To establish a link between reductions in APUM24 expression and the phenotype of the apum24-2 mutant, we produced APUM24-targeted artificial microRNA (amiRNA) lines. Two of these lines (apum24i-1 and apum24i-2), in which APUM24 expression was repressed at a level comparable to that in the apum24-2 mutant (Supplemental Figure 1A), similarly developed seeds with abnormal shapes (Supplemental Figure 1B). These results suggest that reductions in APUM24 expression affect embryogenesis.
Interactions of APUM24 with Pre-rRNA Processing-Associated Proteins in the Nucleolus
To reveal the biological process in which APUM24 is involved, we performed proteomic identification of proteins that coimmunoprecipitated with APUM24. Using cell lysates of wild-type Arabidopsis T87 cells and cells expressing MYC-tagged APUM24, we identified candidate proteins that specifically interact with APUM24. The proteins that were detected only in cells expressing MYC-tagged APUM24 included many RPs and several pre-rRNA processing factors for 60S ribosome biogenesis (Kojima et al., 2007; Weis et al., 2015b; Burgess et al., 2015) (Table 2). Furthermore, coexpression analysis using the ATTEDII program (http://atted.jp; Obayashi et al., 2007) suggested that APUM24 is coexpressed with BIOGENESIS OF RIBOSOMES IN XENOPUS1-2 (BRX1-2) (Supplemental Figure 2), whose product coimmunoprecipitated with APUM24 (Table 2). BRX1-2 and its paralog, BRX1-1, belong to the BRIX domain-containing protein family, which might function in ribosome biogenesis in animals, yeast, and plants (Kaser et al., 2001; Weis et al., 2015b). Thus, APUM24 appears to function in ribosome biogenesis.
Table 2. Proteins That Were Specifically Coimmunoprecipitated with MYC-Tagged APUM24.
| Scorea | Protein Name | AGI Code | Distinct Peptides | %AA Coverageb | Total Spectral Intensity |
|---|---|---|---|---|---|
| Pre-rRNA processing-related proteins | |||||
| 158.97 | Homolog of NOP56 | AT1G56110 | 10 | 22.0 | 1.88E+08 |
| 72.31 | Homolog of NOP58 | AT3G05060 | 5 | 10.8 | 3.38E+07 |
| 69.58 | Homolog of NOP58 | AT5G27120 | 4 | 7.5 | 4.33E+07 |
| 33.28 | BRX1-2 | AT1G52930 | 3 | 10.6 | 4.02E+07 |
| 28.56 | OLI2/NOP2A | AT5G55920 | 2 | 2.6 | 5.66E+07 |
| RP | |||||
| 139.8 | RPL4A | AT3G09630 | 8 | 25.3 | 1.24E+08 |
| 74.02 | RPL8A | AT2G18020 | 4 | 20.9 | 6.95E+07 |
| 65.90 | RPL5B | AT5G39740 | 4 | 15.2 | 4.40E+07 |
| 65.32 | RPS13B | AT4G00100 | 4 | 33.7 | 1.62E+08 |
| 60.69 | RPL14B | AT4G27090 | 4 | 34.3 | 2.16E+08 |
| 60.51 | RPL6C | AT1G74050 | 4 | 15.0 | 3.99E+07 |
| 53.47 | RPL3A | AT1G43170 | 4 | 9.2 | 1.28E+07 |
| 49.47 | RPL7B | AT2G01250 | 4 | 19.0 | 1.55E+08 |
| 47.21 | RPL23Aa | AT2G39460 | 3 | 22.0 | 4.44E+07 |
| 46.39 | RPL13aB | AT3G24830 | 3 | 14.5 | 1.22E+08 |
| 46.15 | RPS18A | AT1G22780 | 3 | 15.7 | 2.88E+07 |
| 41.23 | RPL12B | AT3G53430 | 3 | 22.2 | 2.04E+08 |
| 40.98 | RPP0B | AT3G09200 | 3 | 10.3 | 2.18E+07 |
| 35.16 | RPL90B | AT1G33120 | 3 | 13.9 | 6.04E+08 |
| 33.83 | RPS15Aa | AT1G07770 | 3 | 26.1 | 1.55E+07 |
| 30.41 | RPL7A | AT1G80750 | 3 | 10.5 | 3.98E+07 |
| 28.11 | RPL27C | AT4G15000 | 3 | 14.0 | 2.11E+09 |
| 27.53 | RPL7aBc | AT3G62870 | 3 | 6.2 | 1.42E+07 |
| 26.86 | RPL7aAc | AT2G47610 | 3 | 6.2 | 1.09E+07 |
| 26.02 | RPS14B | AT3G11510 | 3 | 14.6 | 2.79E+07 |
| 23.07 | RPL18C | AT5G27850 | 1 | 5.8 | 2.55E+07 |
| 22.77 | RPL11B | AT3G58700 | 1 | 7.6 | 2.12E+07 |
| 21.95 | RPS6A | AT4G31700 | 1 | 6.0 | 1.09E+07 |
| 21.93 | RPL15A | AT4G16720 | 2 | 10.2 | 2.24E+07 |
| Others | |||||
| 57.73 | ADL1E | AT3G60190 | 4 | 9.7 | 5.13E+07 |
| 48.84 | Unknown protein | AT5G57120 | 3 | 11.5 | 1.30E+07 |
| 45.60 | Nhp2-like | AT5G08180 | 3 | 14.7 | 6.91E+07 |
| 37.06 | ACT2 | AT3G18780 | 3 | 5.5 | 4.85E+06 |
| 35.52 | HSC70-2 | AT5G02490 | 3 | 4.1 | 1.58E+08 |
| 32.33 | DCP1 | AT1G08370 | 3 | 5.4 | 6.96E+07 |
| 30.27 | HTA7 | AT5G27670 | 3 | 21.3 | 1.00E+08 |
| 27.62 | Unknown protein | AT5G09840 | 3 | 4.0 | 2.60E+08 |
| 23.93 | MAK16 protein-related | AT1G23280 | 1 | 4.2 | 3.86E+06 |
| 22.51 | Unknown protein | AT3G18600 | 2 | 4.5 | 1.69E+07 |
Calculated by Spectrum Mill (Agilent).
AA, amino acid.
Indistinguishable.
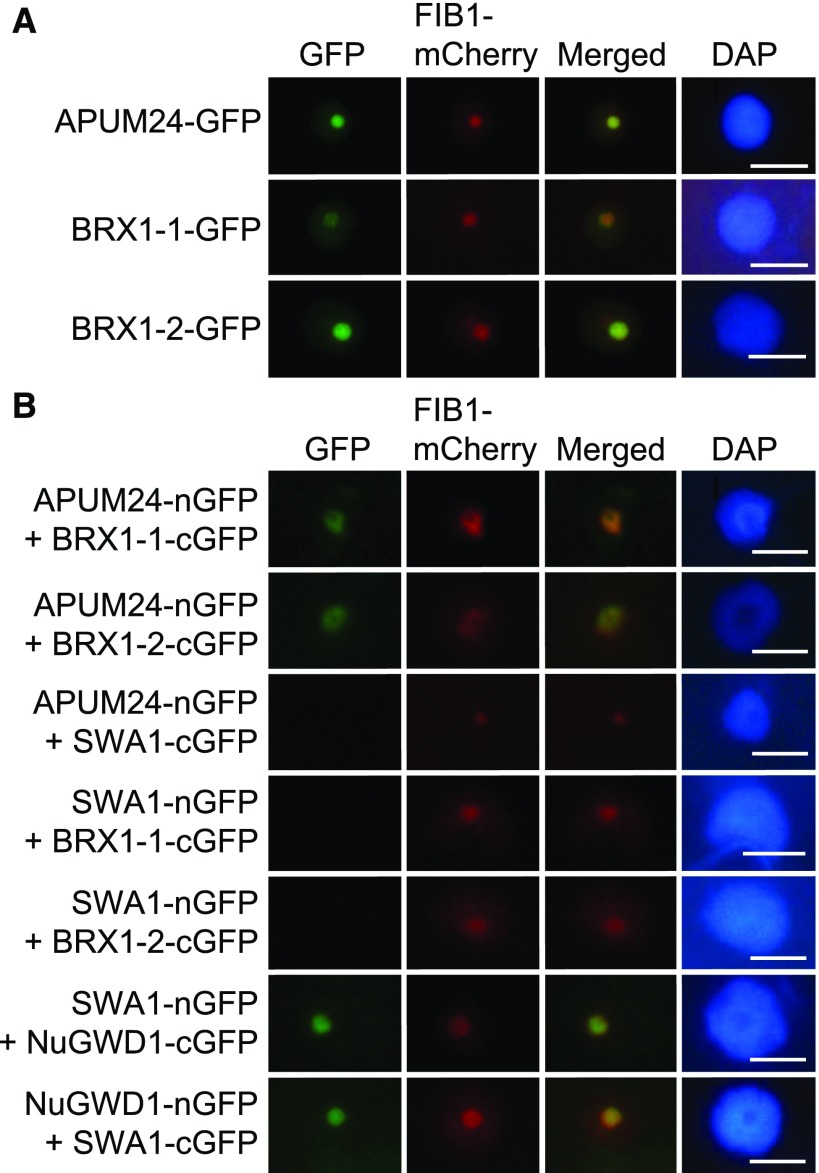
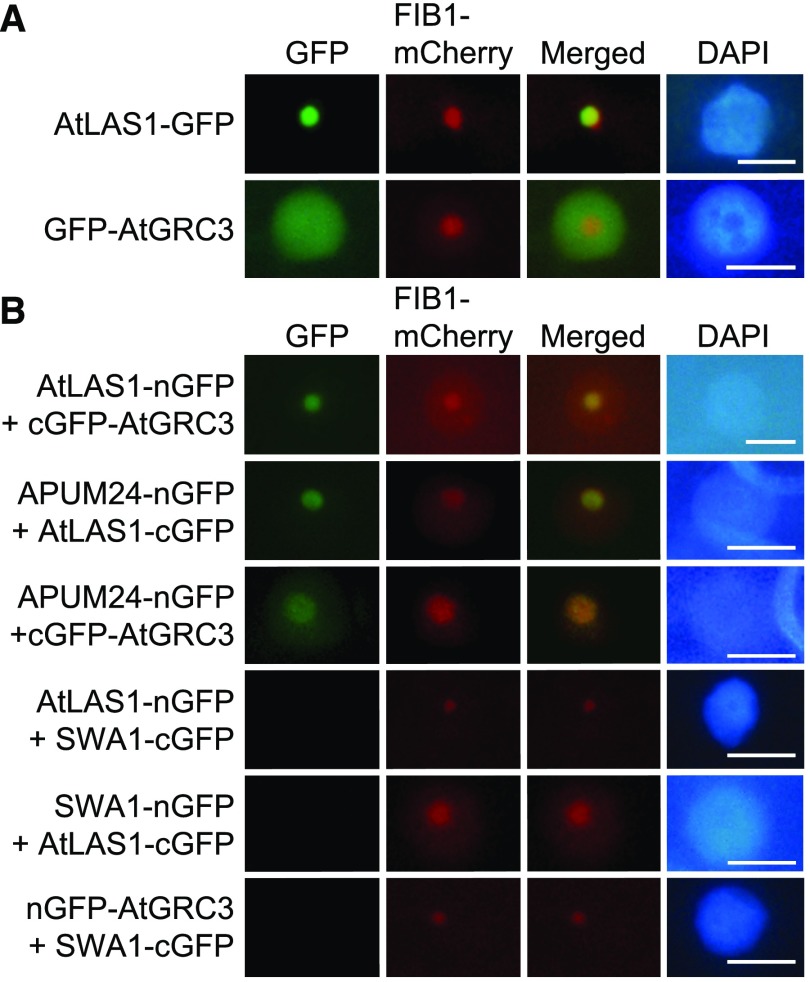
To investigate the interactions of APUM24 with BRX1-1 and BRX1-2 in vivo, we performed bimolecular fluorescence complementation (BiFC) analysis with the N-terminal half of GFP (nGFP) and the C-terminal half of GFP (cGFP) (Walter et al., 2004). Prior to BiFC analysis, we confirmed the nucleolar localization of APUM24 by coexpressing it with mCherry fused to fibrillarin1 (FIB1-mCherry), a nucleolus-localized marker protein (Barneche et al., 2000; Tam et al., 2010; Figure 2A). As reported previously (Weis et al., 2015b), BRX1-1 and BRX1-2 also localized to the nucleolus in our assay (Figure 2A). We observed GFP fluorescence in the nucleolus after transient coexpression of the APUM24-nGFP fusion protein with cGFP fused to BRX1-1 or BRX1-2 in Nicotiana benthamiana leaves (Figure 2B). In this assay, we also coexpressed FIB1-mCherry to identify transformed cells and to visualize the nucleolus. Furthermore, we used another nucleolar localized protein, SLOW WALKER1 (SWA1), a homolog of U3 small nucleolar ribonucleoprotein15 of the yeast 18S rRNA processing complex (Shi et al., 2005), as a negative control (Figure 2B). Because GFP fluorescence was observed in the nucleolus after coexpression of SWA1-nGFP with NuGWD1-cGFP and NuGWD1-nGFP with SWA1-cGFP (Figure 2B), as reported previously (Ishida et al., 2016), we considered SWA1-nGFP and SWA1-cGFP as appropriate negative controls in this experiment. Neither the coexpression of SWA1-cGFP with APUM24-nGFP nor the coexpression of SWA1-nGFP with BRX1-1-cGFP or BRX1-2-cGFP (Figure 2B) resulted in GFP fluorescence. Furthermore, GFP fluorescence was not detected in cells solely expressing nGFP or cGFP fused to APUM24, BRX1-1, BRX1-2, SWA1, and NuGWD1 (Supplemental Figure 3). Thus, the results of BiFC analysis suggested that APUM24 interacts with pre-rRNA processing-associated factors in vivo.
Figure 2.
Interactions of APUM24 with BRX1-1 and BRX1-2 in the Nucleolus in Vivo.
(A) Nucleolar localization of GFP fused to APUM24, BRX1-1, or BRX1-2 (APUM24-GFP, BRX1-1-GFP, or BRX1-2-GFP). Green fluorescence from GFP and fluorescence from FIB1-mCherry as a nucleolar marker were simultaneously monitored by transiently expressing these proteins in N. benthamiana leaves. Images for GFP were merged with those for fluorescence from mCherry (Merged). The 4′,6-diamidino-2-phenylindole (DAPI) staining indicates nucleoplasm. Bars = 10 μm.
(B) Representatives of BiFC analyses. Typical images obtained during the observation of many transformed cells are shown. APUM24 fused to nGFP (APUM24-nGFP) was coexpressed with cGFP-fused BRX1-1 and BRX1-2 (BRX1-1-cGFP and BRX1-2-cGFP) in N. benthamiana leaves. Combinations of APUM24-nGFP and SWA1-cGFP, and SWA1-nGFP and BRX1-1-cGFP or BRX1-1-cGFP served as negative controls. The combination of SWA1-nGFP and NuGWD1-cGFP, and NuGWD1-nGFP and SWA1-cGFP served as controls for the production of active SWA1-nGFP and SWA1-cGFP proteins. FIB1-mCherry and DAPI staining are the same as in (A). Bars = 10 μm.
Pleiotropic Defects in Developmental Processes in apum24-2 mutants
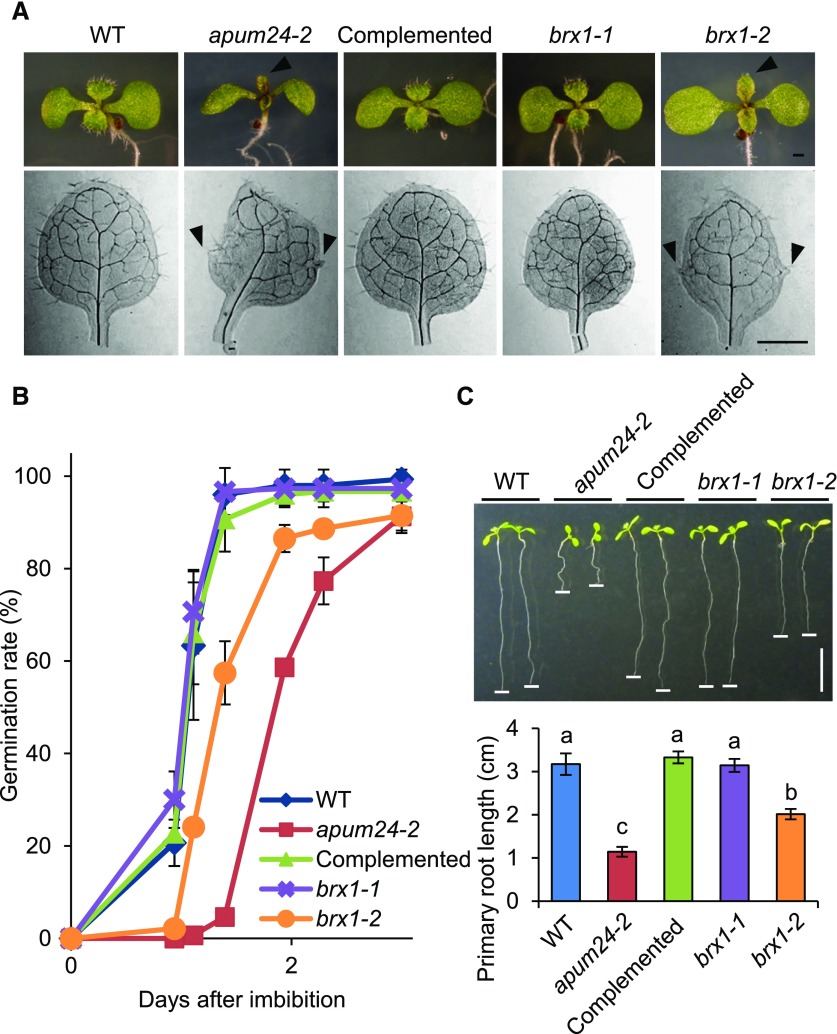
Arabidopsis RP and RBF mutants frequently exhibit delayed embryogenesis and germination and short primary roots, as well as abnormal alignment of vascular bundle tissues, leading to the formation of leaves with abnormal shapes, such as pointed and/or serrated leaves (Byrne, 2009; Horiguchi et al., 2012; Weis et al., 2015a). We therefore investigated whether the apum24-2 mutant, as well as brx1-1 and brx1-2 (Weis et al., 2015b), displays such phenotypes. Like the brx1-2 mutant, which has pointed and serrated true leaves (Weis et al., 2015b), apum24-2 had abnormally shaped true leaves (Figure 3A). Furthermore, germination was delayed in both apum24-2 and brx1-2 (Figure 3B), and their primary roots were shorter than those of the wild type (Figure 3C). Similar to the apum24-2 mutant, two independent APUM24-targeted amiRNA lines also showed pointed true leaves with serrated edges and root growth defects (Supplemental Figures 1C to 1E). Under other conditions, we observed that the brx1-1 mutation caused pointed- and serrated-leaf phenotypes (Supplemental Figure 4), as mentioned in the Discussion. Because some Arabidopsis mutants defective in ribosome biogenesis or function showed antibiotic resistance (Abbasi et al., 2010; Rosado et al., 2010), we also investigated primary root growth of apum24-2 in the presence of two antibiotic compounds (streptomycin and chloramphenicol) and found that these antibiotics had only a small effect on root elongation in the apum24-2 mutant (Supplemental Figure 5). These results imply that the nucleolus-localized protein APUM24 is involved in the control of development via its activity to regulate ribosome biogenesis.
Figure 3.
apum24-2 and brx1-2 Mutants Have Similar Phenotypes.
(A) Photographs of shoots of 7-d-old seedlings (upper panel) and true leaves of 10-d-old seedlings (lower panel) of wild-type Arabidopsis, apum24-2, and the complemented line (Complemented), as well as the brx1-1 and brx1-2 mutants. Arrowheads indicate pointed leaves (upper panel) and serrated edge (lower panel). Bars = 1 mm.
(B) Time course of germination rates. The germination rates of 50 seeds were counted over time. Error bars indicate sd (n = 3).
(C) Photographs (upper panel) and quantification (lower panel) of primary root lengths of 7-d-old seedlings. Horizontal white lines were put on the tip of the primary roots. Error bars indicate sd (n = 7). Statistical significance was determined by ANOVA, followed by a Tukey-Kramer test. Means that are significantly different from each other (P < 0.05) are indicated by different letters. Bar = 1 cm.
The APUM24-Containing Complex Is Involved in 45S Pre-rRNA Processing
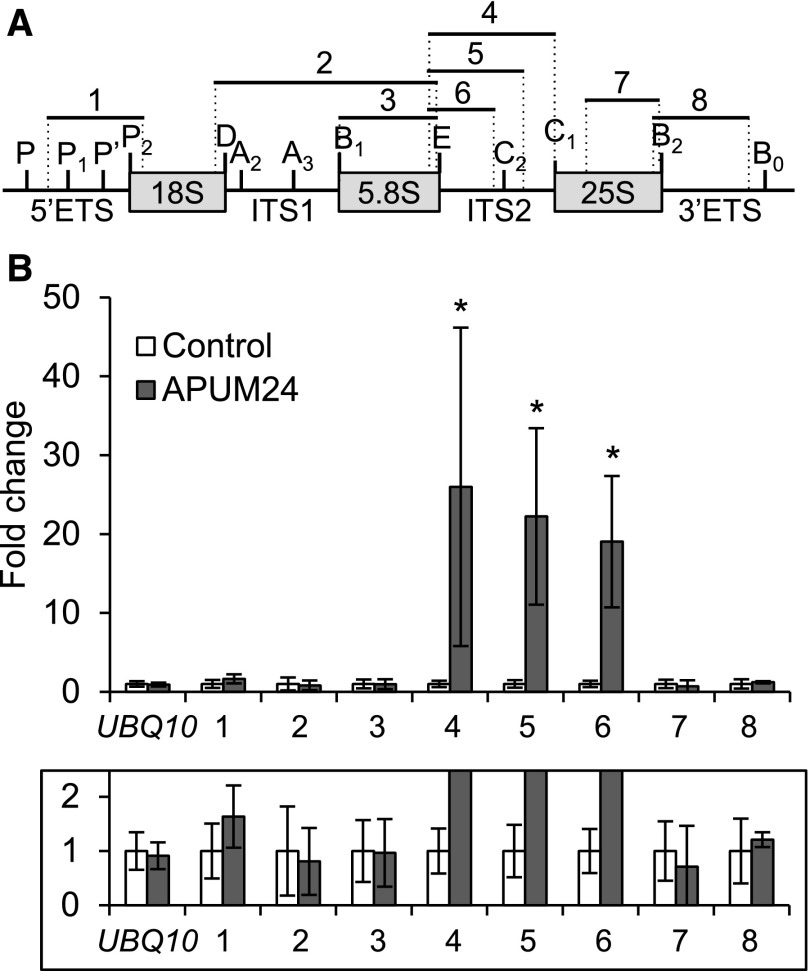
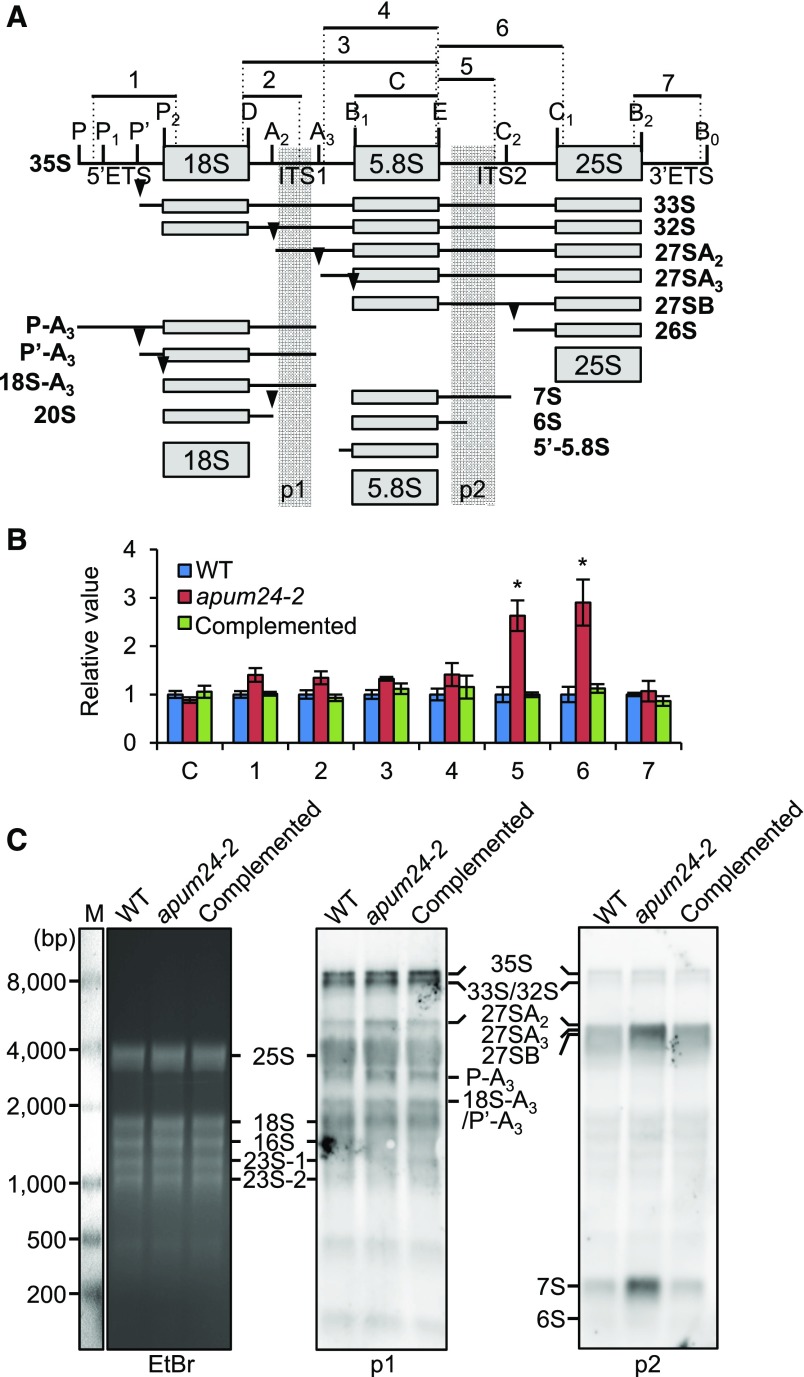
Since APUM24 has a putative RNA binding domain, PUM-HD (Tam et al., 2010), and apum24-2 mutant showed ribosome-related developmental phenotypes, we performed RNA immunoprecipitation (RIP) assays to examine the binding of the APUM24-contaning complex to pre-rRNA. We prepared cell lysates of apum24-2 expressing (or not expressing) FLAG-tagged APUM24 under the control of its own promoter and performed immunoprecipitation with anti-FLAG antibodies after cross-linking proteins to RNA using formaldehyde and sonication to fragment RNA. After confirming immunoprecipitation of FLAG-tagged APUM24 (Supplemental Figure 6), we quantified the levels of rRNA that coimmunoprecipitated with APUM24 by performing RT-qPCR with several pairs of primers used to amplify different regions of the pre-rRNA or mature rRNA (Figure 4A). Among various fragments produced from the pre-rRNA and mature rRNA, only fragments that contained a part of ITS2 (primer sets 4, 5, and 6) were significantly enriched, depending on the expression of FLAG-tagged APUM24 (Figure 4B). These results suggest that the APUM24-containing complex specifically interacts with ITS2-containing pre-rRNA intermediates (see Figure 5A).
Figure 4.
RIP Assay for the Binding of APUM24 to 45S Pre-rRNA.
(A) Organization of 45S pre-rRNA and positions of specific primers used to amplify regions 1 to 8. Vertical lines indicate the site of endo- or exonuclease processing steps. ETS and ITS are external and internal transcribed spacer, respectively.
(B) RT-qPCR analysis of RNA coimmunoprecipitated with APUM24-FLAG protein by anti-FLAG antibodies. Enrichment of pre-rRNA or rRNA with specific regions (regions 1 to 8 in [A]) by coimmunoprecipitation was calculated with values obtained using the output sample versus those using the input sample. UBQ10 is shown as a non binding control. Error bars represent sd (n = 3). Asterisks indicate statistically significant differences between values obtained with apum24-2 (Control) and apum24-2 expressing APUM24-FLAG (APUM24) by Student’s t test (P < 0.05). The lower panel shows a view enlarged in the vertical direction.
Figure 5.
Pre-rRNA Processing in the apum24-2 Mutant.
(A) Diagram illustrating the major pathways of 35S pre-rRNA processing and 35S pre-rRNA processing intermediates in plants (Weis et al., 2015a, 2015b). Vertical lines and arrowheads indicate the sites of endo- or exonuclease processing. ETS and ITS are the external transcribed spacer and the internal transcribed spacer, respectively. Regions 1 to 7 indicate pre-rRNA fragments that were amplified by PCR with specific primer sets to quantify the respective pre-rRNAs; 5.8S rRNA (region C) was also amplified to compare the total amounts of total rRNA in each sample in (B). Regions p1 and p2 indicate the positions of probes for RNA gel blot detection in (C).
(B) Relative levels of processing intermediates in the apum24-2 mutant and the complemented apum24-2 line grown in the presence of 150 mM glucose. Regions C and 1 to 7 are indicated in (A). The values are expressed relative to the levels in wild-type seedlings, and UBQ10 was used as an internal control to normalize the values. Error bars represent sd (n = 3). Asterisks indicate statistically significant differences between the mutants and the wild type by Student’s t test (P < 0.05).
(C) RNA gel blot detection of pre-rRNA intermediates with probe p1 or p2 whose positions are indicated in (A). The ethidium bromide (EtBr)-stained gel image is shown as a loading control. Detected pre-rRNA intermediates are indicated. Abundant rRNAs (25S, 18S, 16S, and 23S rRNAs) were nonspecifically detected. M, RNA marker.
Next, we monitored the rRNA processing steps in the apum24-2 mutant by RT-qPCR to detect and quantify processing intermediates. We used a primer for the sequence that would remain in rRNA and a primer for 5′ETS, 3′ETS, ITS1, or ITS2 in each PCR to quantify respective processing intermediates, whereas primers for the 5.8S rRNA sequence were used to quantify the total amounts of rRNA and pre-rRNA (Figure 5A). The results indicate that intermediates containing ITS2 (i.e., intermediates containing region 5 or 6) accumulated in the apum24-2 mutant in excess, although the amounts of other processing intermediates as well as the total amounts of rRNA in the apum24-2 mutant were similar to those in the wild type (Figure 5B). As shown in Figure 5A, it is known that there are multiple forms of ITS2-containing intermediates (Weis et al., 2015a, 2015b). Thus, we performed RNA gel blot analysis to characterize further the pre-rRNA processing intermediates in apum24-2. Using probe p1 designed to detect 35S, 33S/32S, 27SA2, P-A3, and 18S-A3/P’-A3 pre-rRNAs (Figure 5A), we detected bands corresponding to these pre-rRNAs, although mature rRNAs were also weakly detected, probably because of nonspecific interactions between the probe DNA and the very abundant mature rRNAs. Very similar band patterns were obtained using RNA from the wild-type and apum24-2 Arabidopsis plants when using probe p1 (Figure 5C). On the other hand, consistent with the results of RT-PCR quantification of pre-rRNA (Figure 5B), using the probe p2 designed to detect 35S, 33S/32S, 27SA2, 27SA3, 27SB, 7S, and 6S pre-rRNAs, we detected bands corresponding to all of these pre-rRNAs but with overrepresentation of 27SA3 and/or 27SB pre-rRNA and 7S pre-rRNA bands, indicating that these RNAs accumulated in the apum24-2 mutant (Figure 5C)
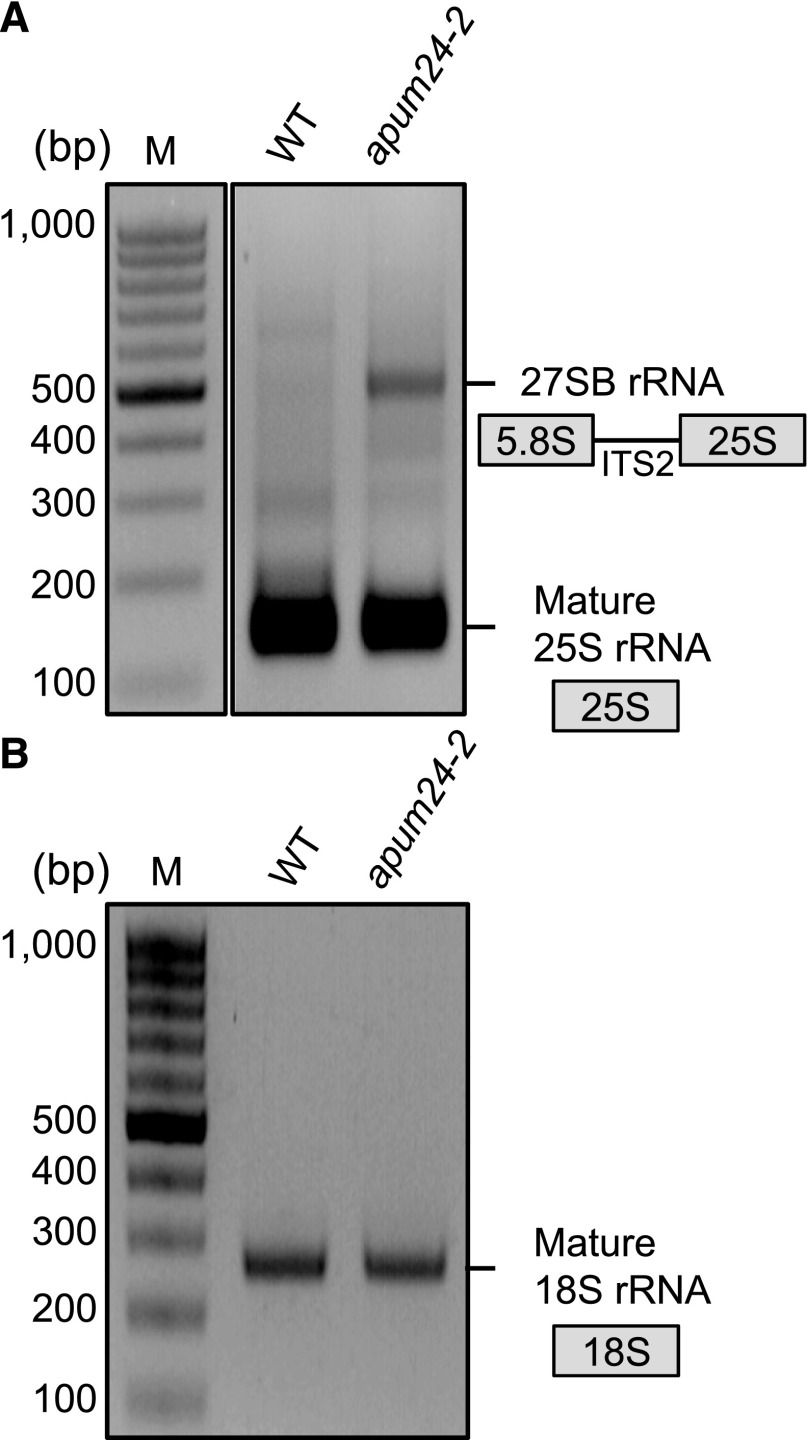
To investigate which pre-rRNA, 27SA3 or 27SB pre-rRNA, overaccumulated in the apum24-2 mutant, we subjected total RNA to circular RT-PCR (cRT-PCR), which identifies the 5′ and 3′ ends of RNA via self-circularization with T4 RNA ligase (Abbasi et al., 2010). Total RNA extracted from the wild-type and apum24-2 plants was circularized and reverse transcribed, and PCRs were performed with primers designated to amplify the sequences attached to the 25S or 18S rRNA sequences (see details in Methods). In the amplification targeting the 25S rRNA sequence, we detected a PCR product specific to RNA from the apum24-2 mutant in addition to a PCR product corresponding to mature 25S rRNA (Figure 6A). Sequencing revealed that this product originated from 27SB pre-rRNA, not from 27SA3 pre-rRNA. It is also noteworthy that we did not detect the PCR product of 26S pre-rRNA that contains a part of ITS2 (Figure 5A). Additionally, we did not detect any unprocessed intermediates through amplification targeting the 18S rRNA sequence (Figure 6B). Taken together, the results of RT-PCR-based pre-rRNA quantification, RNA gel blot analysis, and cRT-PCR analysis suggested that 27SB and 7S pre-rRNAs accumulated in excess in the apum24-2 mutant.
Figure 6.
Circular RT-PCR Analysis Revealed Overaccumulation of 27SB Pre-rRNA in the apum24-2 Mutant.
Products of cRT-PCR targeting RNA containing the 25S rRNA sequence (A) or 18S rRNA sequence (B) were analyzed by 2% agarose gel electrophoresis and the gels were stained with ethidium bromide. Three independent clones corresponding to ∼150- and 500-bp products in (A) were sequenced to confirm that the former and latter bands were derived from mature 25S rRNA and 27SB rRNA, respectively. Three independent clones corresponding to the ∼250-bp product in (B) were also sequenced to confirm that it was derived from mature 18S rRNA. M, DNA marker.
APUM24 Interacts with the Putative ITS2 Endonuclease Complex in the Nucleolus
Recently, Gasse et al. (2015) identified the ITS2 processing complex in the nucleolus of Saccharomyces cerevisiae. This complex contains ScLas1, which cleaves pre-rRNA at the C2 site, and ScGrc3, which phosphorylates the 5′ end of the cleaved site, as well as ScRai1 and ScRat1, which possess exonuclease activities for the generation of mature 25S rRNA. By performing BLAST searches, we found that At5g12220 and At5g11010 are putative orthologs of ScLas1 and ScGrc3, respectively (Supplemental Table 1 and Supplemental Figures 7 and 8). Although the functions of their products have not yet been clarified, we designated At5g12220 and At5g11010 as AtLAS1 and AtGRC3, respectively. Although CLPS3 and CLPS5 are similar to ScGRC3 (Supplemental Table 1), they have been already characterized as homologs of the human polyadenylation factor CLP1 (Xing et al., 2008). Thus, they were excluded from further analyses.
Analysis with the ATTEDII program suggested that AtLAS1 and AtGRC3 are coexpressed with RBF genes and APUM24 (Supplemental Figure 9). Furthermore, AtLAS1 and AtGRC3 fused to GFP localized to the nucleolus (Figure 7A), although GFP-fused AtGRC3 was detected throughout the nucleus. We therefore examined the interactions of APUM24 with AtLAS1 and AtGRC3 via BiFC assays. The GFP fluorescence patterns detected in these assays revealed interactions of APUM24 with AtLAS1 and AtGRC3, as well as interaction of AtLAS1 and AtGRC3, in the nucleolus (Figure 7B). We did not detect GFP fluorescence using the SWA1 negative control proteins that were used in Figure 2 and also confirmed that cells expressing only nGFP or cGFP fused to AtLAS1 or AtGRC3 alone did not emit fluorescence (Supplemental Figure 3). Together, the results suggested that APUM24, together with AtLAS1 and AtGRC3, is likely involved in ITS2 removal.
Figure 7.
Interactions of APUM24 with AtLAS1 and AtGRC3 in the Nucleolus.
(A) Subcellular localization of AtLAS1 (At5g12220.1)-GFP and GFP-AtGRC3 (At5g11010.2) fusion proteins. Green fluorescence from AtLAS1-GFP and GFP-AtGRC3 transiently expressed in N. benthamiana leaves (GFP) and fluorescence from FIB1-mCherry that was simultaneously expressed as a nucleolar marker protein (mCherry) are shown. GFP images were merged with mCherry images (Merged). DAPI staining indicates nucleoplasm. Bars = 10 μm.
(B) Typical images obtained by BiFC analyses. AtLAS1-nGFP was coexpressed with cGFP-AtGRC3, and APUM24-nGFP was coexpressed with AtLAS1-cGFP or cGFP-AtGRC3 in N. benthamiana leaves. The combinations of AtLAS1-nGFP, AtLAS1-cGFP, or nGFP-AtGRC3 with SWA1-cGFP or SWA1-nGFP are shown as negative controls. FIB1-mCherry and DAPI staining are the same as in (A). Bars = 10 μm.
The apum24-2 and brx1-2 Mutants Are Hypersensitive to High Concentrations of Sugar
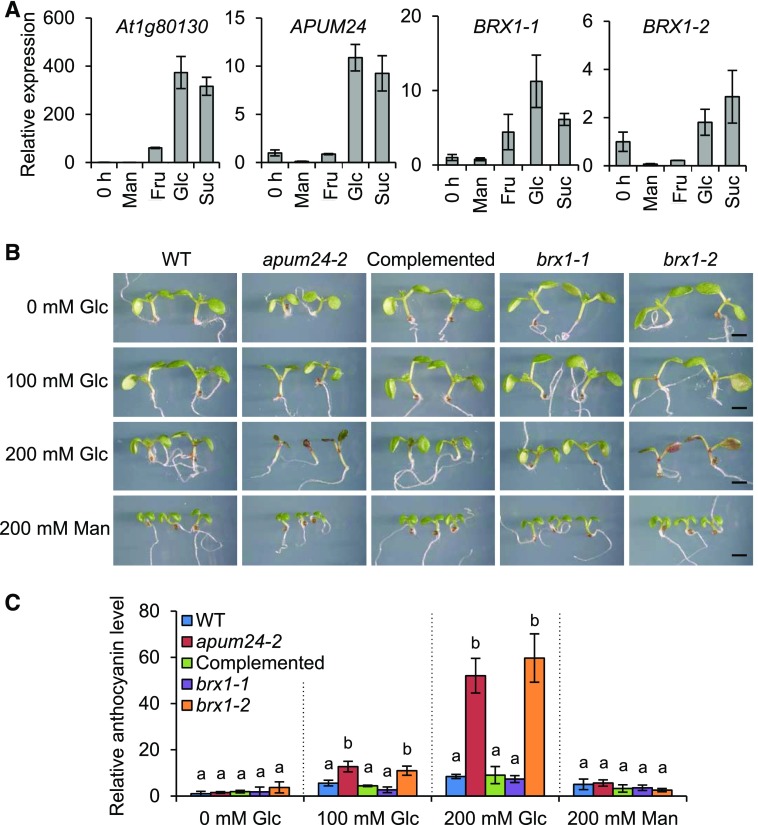
The transcription of ribosome biogenesis- or function-related genes and ribosomal DNA is generally activated by sugar provision, and APUM24 is a sugar-inducible gene (Aki and Yanagisawa, 2009; Kojima et al., 2007; Ishida et al., 2016). Furthermore, we confirmed the sugar-inducible expression of BRX1-1 and BRX1-2, as well as APUM24, using wild-type seedlings treated with various metabolizable sugars (glucose, sucrose, and fructose), with nonmetabolizable sugar (mannitol) serving as an osmotic control (Figure 8A). We therefore investigated the responses of apum24-2, brx1-1, and brx1-2 to sugar by monitoring plant growth on medium containing 0, 100, or 200 mM glucose or 200 mM mannitol (Figures 8B and 8C). High concentrations of metabolizable sugar repress cotyledon expansion and promote the accumulation of anthocyanin, a sugar stress marker, in Arabidopsis seedlings (Martin et al., 2002). These effects were much more evident in apum24-2 and brx1-2 than in the wild-type plants, indicating that impairing the functions of APUM24 and BRX1-2 causes Arabidopsis seedlings to become hypersensitive to high concentrations of sugar.
Figure 8.
Sugar Responses of apum24-2, brx1-1, and brx1-2.
(A) Relative expression levels of APUM24, BRX1-1, and BRX1-2 in 8-d-old seedlings treated with 50 mM mannitol (Man), fructose (Fru), glucose (Glc), or sucrose (Suc) for 3 h. The expression levels of these genes in untreated seedlings (0 h) were set to 1. The sugar-responsive marker gene, At1g80130 (Price et al., 2004), was used as a control, and UBQ10 was used as an internal control. Error bars represent sd (n = 3).
(B) Seedlings of the wild type, apum24-2, the complemented apum24-2 line (Complemented), brx1-1, and brx1-2 were grown for 7 d on 1/2 MS medium plates containing the indicated concentrations of glucose (Glc) or mannitol (Man). Bar = 1 mm.
(C) Relative anthocyanin levels in the seedlings in (B). The anthocyanin content of wild-type seedlings grown without sugar was set to 1. Error bars represent sd (n = 3). Statistical significance was determined by ANOVA, followed by a Tukey-Kramer test under each experimental condition. Means that are significantly different from each other (P < 0.05) are indicated by different letters.
Nucleolar Stress Occurs in Response to the Sugar Status of apum24-2
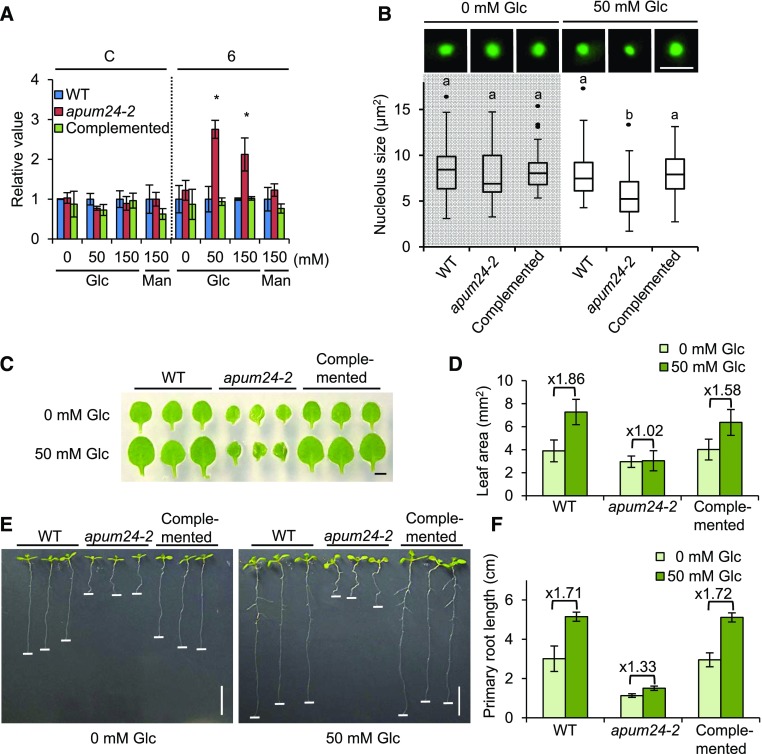
To investigate the link between impaired rRNA synthesis and the modified sugar response, we compared the levels of rRNA processing intermediates in apum24-2 seedlings grown on medium containing different concentrations of glucose (0, 50, and 150 mM) or 150 mM mannitol (Figure 9A). Interestingly, unprocessed intermediates containing ITS2 overaccumulated specifically in the presence of exogenous glucose, indicating that the overaccumulation of rRNA processing intermediates depends on the sugar status of the cell.
Figure 9.
Sugar-Dependent Nucleolar Stress in the apum24-2 Mutant.
(A) Sugar-dependent accumulation of the pre-rRNA processing intermediates in the apum24-2 mutant. Total RNA was prepared from seedlings of the wild type, apum24-2, and the complemented line (Complemented) grown on medium containing different concentrations of glucose (Glc) (0, 50, and 150 mM) or 150 mM mannitol (Man) for 7 d. For RT-qPCR, primers for amplification of regions 6 and C (indicated in Figure 5A) were used. Error bars represent sd (n = 3). The values are relative to the levels in wild-type seedlings, and UBQ10 was used as an internal control to normalize the values.
(B) Upper panels display representative nucleoli in mesophyll protoplasts from the wild type, apum24-2, and the complemented apum24-2 line (Complemented) grown on medium containing 0 or 50 mM glucose (Glc) for 7 d. The nucleolus was visualized by observing green fluorescence from transiently expressed FIB1-GFP. The graph shows a box plot of the quantification of nucleolus size (n = 50). Dots indicate outliers. Statistical significance was determined by ANOVA, followed by a Tukey-Kramer test under each experimental condition. Means that are significantly different from each other (P < 0.05) are indicated by different letters. Bar = 5 μm.
(C) and (D) Representatives (C) and area quantification (D) of the first true leaves of the plants grown on medium containing 0 or 50 mM glucose (Glc) for 9 d, respectively. Bar = 1 mm. Error bars represent sd (n = 20).
(E) and (F) Representatives (E) and quantification of the primary root length (F) of the plants grown on medium containing 0 or 50 mM glucose (Glc) for 9 d. Error bars represent sd (n = 9). Horizontal white lines were placed on the tips of the primary roots. Bar = 1 cm.
In yeast and animal cells, the accumulation of abnormal rRNA due to dysfunctional rRNA processing causes nucleolar stress, which is characterized by reduced or enlarged nucleolar size and altered cell growth/proliferation and stress responses (Holmberg Olausson et al., 2012; Zhou et al., 2015). In plants, the ribosome biogenesis-related DEAD-box helicase RH10 acts as a link between temperature-dependent rRNA processing defects and nucleolus enlargement (Matsumura et al., 2016). Thus, we investigated whether the sugar-dependent overaccumulation of pre-rRNA intermediates causes nucleolar stress in the apum24-2 mutant. We transiently expressed FIB1 fused to GFP as a nucleolar marker protein in Arabidopsis mesophyll protoplasts prepared from leaves of wild-type and apum24-2 seedlings grown on 0 or 50 mM glucose-containing medium. Since the nucleolus in these cells could be visualized by observing green fluorescence, we measured the sizes of nucleolar zones and evaluated the effects of the apum24-2 allele and glucose on nucleolar size (Figure 9B). The nucleolar sizes in wild-type cells were comparable regardless of exogenous glucose application. Moreover, no difference was observed in the size of the nucleolus in protoplasts from wild-type versus apum24-2 seedlings grown without exogenous glucose. However, in protoplasts from seedlings grown in the presence of 50 mM glucose, the nucleolus size was significantly smaller in the apum24-2 mutant than in the wild type (Figure 9B). To further confirm this observation, we repeated the experiment with GFP fused to another nucleolus-localized protein, SWA1, that did not interact with APUM24 (Figure 2B) and obtained similar results (Supplemental Figure 10A). These results indicate that nucleolar stress is dependent on sugar status in the apum24-2 mutant. This finding is consistent with the sugar-dependent overaccumulation of processing intermediates containing ITS2 in apum24-2 (Figure 9A) and the hypersensitivity of this mutant to high concentrations of sugar (Figures 8B and 8C).
According to previous reports, nucleolar stress negatively affects cell growth and cell proliferation in mammal and yeast cells (Holmberg Olausson et al., 2012; Zhou et al., 2015). We therefore evaluated the effects of nucleolar stress on cell growth and cell proliferation in the apum24-2 mutant by measuring the sizes of first true leaves of the seedlings grown on medium containing 0 or 50 mM glucose (Figures 9C and 9D). The leaf sizes of the wild-type plants grown in the presence of a low concentration of glucose (50 mM) were larger than those grown in the absence of exogenously supplied glucose. However, interestingly, the apum24-2 mutant did not show a glucose-induced increase in leaf size (Figures 9C and 9D). Similarly, the glucose-induced positive effect on root growth was much lower in the apum24-2 mutant than in the wild type (Figures 9E and 9F). The sugar-induced increases in leaf and root growth of the seedlings of the complemented line were similar to those of wild-type seedlings. Taken together, the results suggest that reductions in APUM24 expression result in a sugar-dependent pre-rRNA processing defect that triggers nucleolus stress and abnormal sugar responses.
DISCUSSION
Our results indicate that APUM24 is an essential gene encoding a novel factor involved in pre-rRNA processing. The apum24-1 null mutant is embryonic lethal, whereas the apum24-2 knockdown mutant displays phenotypic features of ribosome biogenesis or function-defective mutants, suggesting that APUM24 plays a unique role in Arabidopsis. Importantly, the reduced expression of APUM24 induced sugar-dependent defects in pre-rRNA processing and nucleolar stress, ultimately leading to hypersensitivity to high concentrations of sugar and decreases in growth in response to low concentrations of sugar in apum24-2 seedlings. Hence, our findings uncover linkages among ribosome biogenesis, nucleolar stress, and sugar responses in plants, paving the way for the new research field of nucleolar stress signaling and its connection to the regulation of plant growth.
APUM24 Is an Essential Factor with a Unique Role in Pre-rRNA Processing in Arabidopsis
The null mutation of APUM24 led to embryonic lethality, as it influenced both gametogenesis and embryogenesis (Figure 1, Table 1). This finding indicates that APUM24 plays an essential role in pre-rRNA processing. Another APUM protein, APUM23, also functions in pre-rRNA processing (Abbasi et al., 2010; Huang et al., 2014). Indeed, apum23 and apum24-2 mutants exhibit similar phenotypes, namely, abnormal shoot and root development, and defects in pre-rRNA processing (Abbasi et al., 2010; Figures 3 and 5). However, the APUM24-containing complex interacted with ITS2 (Figure 4), while APUM23 interacts with 18S rRNA (Zhang and Muench, 2015). Therefore, the functions of APUM23 and APUM24 are not redundant. The observation that the apum24-1 single mutation led to embryo lethality (Figure 1) strongly supports the unique role of APUM24 in Arabidopsis.
Puf-A (also known as KIAA0020) and Puf6 are the closest proteins to APUM24 among human and yeast proteins, respectively, sharing 74% and 70% similarity with APUM24, respectively. Despite their similarity, the function of Puf-A in ribosome biogenesis is currently unknown. Puf-A is encoded by a gene that is highly expressed in human breast cancer cells and plays a role in the cellular response to genotoxic stress through direct binding to poly(ADP-ribose) polymerase1 to inhibit the activity of this enzyme (Chang et al., 2011; Fan et al., 2013). By contrast, yeast Puf6 plays two roles: One is related to the asymmetric distribution of ASYMMETRIC SYNTHESIS OF HO1 mRNA (Gu et al., 2004) and the other is related to the biogenesis of the 60S ribosome. Puf6 is found in the 60S ribosome fraction during sucrose density gradient centrifugation (Lee et al., 2007; Li et al., 2009). Additionally, the Δpuf6 deletion strain of S. cerevisiae shows a slow growth phenotype like that of ribosome biogenesis defective mutants with 35S, 27S, and 7S pre-rRNA processing defects (Lee et al., 2007; Li et al., 2009). Recently, it was reported that Puf6 is involved in ribosome biogenesis via its role in the loading of Rpl43 into the large subunit of the ribosome in concert with LOCALIZATION OF mRNA1 (Loc1) protein (Yang et al., 2016). Since APUM24 coimmunoprecipitates with 60S ribosome biogenesis-related proteins, including homologs of Nop56, Nop58, BRX1-2, and OLI2 (Table 2; Kojima et al., 2007; Weis et al., 2015b; Burgess et al., 2015) and APUM24 is involved in ITS2 removal (Figures 5 and 6), APUM24 and Puf6 may share a conserved role as components of early pre-60S particles. However, no homolog of Loc1 has been found yet in Arabidopsis, and the Δpuf6 deletion strain did not show lethality, in contrast to the Arabidopsis apum24-1 mutant (Table 1). Therefore, APUM24 and Puf6 might also have different roles in plant and yeast cells. Further analyses will be necessary to clarify the relationship between Arabidopsis APUM24 and yeast Puf6.
APUM24 Is Involved in the Removal of ITS2 from Pre-rRNA in Arabidopsis
The processing of 45S pre-rRNA, an essential step for maturation of the ribosome, begins with independent cleavages at several sites by distinct endonucleases (Weis et al., 2015a; Figure 5A). RNA fragments originated from ITS2 were specifically enriched by coimmunoprecipitation with APUM24 in RIP assays (Figure 4), and APUM24 possesses PUM-HD as a putative RNA binding domain. Thus, APUM24 itself might bind to ITS2 directly. Furthermore, processing intermediates, 27SB and 7S rRNAs, accumulated to higher levels in the apum24-2 mutant than in the wild type, while the levels of other processing intermediates were similar in the wild type versus the apum24-2 mutant (Figures 5 and 6). Therefore, APUM24 is likely involved in ITS2 removal. We also demonstrated nucleolar interactions of APUM24 with Arabidopsis proteins homologous to yeast ScLas1 and ScGrc3 (Figure 7B). ScLas1 is a member of the Higher Eukaryotes and Prokaryotes Nucleotide binding (HEPN) endonuclease superfamily (Anantharaman et al., 2013), and the ScLas1 complex, containing ScLas1, ScGrc3, ScRat1, and ScRai1, forms a higher order complex, the RNA processome, for the degradation of ITS2 in pre-rRNA (Gasse et al., 2015). Although we did not examine the enzyme activity of AtLAS1, the RX4H motif (an active site of endo-RNase) (Anantharaman et al., 2013) is conserved in AtLAS1 (Supplemental Figure 7), implying that AtLAS1 might be integrated into an RNA processome, together with APUM24, involved in cleavage at the C2 site in Arabidopsis. Therefore, we assume that APUM24 is involved in ITS2 removal via a C2 cleavage step and not via a degradation step from 7S to 6S pre-rRNA. Although we detected overaccumulation of 7S pre-rRNA in addition to 27SB pre-rRNA, we speculate that this may be a side effect of overaccumulation of 27SB pre-rRNA. The nucleolar surveillance system, 3′-5′ exonucleolytic RNA decay pathways for the quality control of pre-rRNA processing, might be involved in this phenomenon (Lafontaine, 2010).
We demonstrated that APUM24 interacts with BRX1-1 and BRX1-2. These homologs of Xenopus laevis BRX and yeast Brx1 are involved in the biogenesis of the 60S subunit (Kaser et al., 2001), suggesting that APUM24 and BRX1-1/BRX1-2 are integrated into the same protein complex. In a Brx1-depleted yeast strain, cleavage at the A3 site in ITS1 and the subsequent 5′-3′ exonucleolytic trimming of the 5′ end of the cleaved pre-rRNA to produce 5.8S rRNA are defective (see Figure 5A), and intermediates that are not processed at the A3 site accumulate in excess (Kaser et al., 2001). The overaccumulation of processing intermediates containing ITS1 in the Arabidopsis brx1-1 and brx1-2 mutant (Weis et al., 2015b) indicates that, like yeast Brx1, BRX1-1 and BRX1-2 play a role in the removal of ITS1 but not ITS2 (Kaser et al., 2001). These findings suggest that, despite the interaction between APUM24 and BRX1-1/BRX1-2 in the nucleolus, APUM24 and BRX1-1/BRX1-2 are specifically involved in different pre-rRNA processing steps, namely, the removal of ITS1 and ITS2, prompting the question of how the same APUM24- and BRX1-1/BRX1-2-containing complex participates in cleavage at different sites in a specific manner. Although no experimental data exist to resolve this question, we speculate that different isoforms of the protein complex might have different RNA binding specificities. Alternatively, the interaction between APUM24 and BRX1-1/BRX1-2 might be due to the formation of a ternary complex containing an unknown protein, and the complex containing only APUM24 or BRX1-1/BRX1-2 might bind to pre-rRNA. To date, few studies have focused on the molecular mechanism underlying pre-rRNA processing in plants (reviewed in Weis et al., 2015a). Our findings, together with our previous finding that the pre-rRNA processing complex contains plant-specific components (Ishida et al., 2016), emphasize the importance of further analysis of the RNA processome for ribosome biogenesis in plant cells.
Although both brx1-1 and brx1-2 mutants overaccumulate pre-rRNA intermediates, only the brx1-2 mutant displays pointed and serrated leaves (Weis et al., 2015b; Figure 3A). We thus investigated the brx1-1 mutation-dependent phenotype by generating a brx1-1 as2 double mutant because the as2 mutation sometimes potentiates the leaf morphological abnormalities caused by other mutations (Machida et al., 2015). We observed that brx1-1 as2 plants, like brx1-2 plants, developed pointed true leaves with serrated edges, although the as2 mutant itself did not show this phenotype (Supplemental Figures 4A and 4B). Together with our results showing interactions between APUM24 and both BRX1-1 and BRX1-2 (Figure 2), this result supports the previous proposal that the functions of BRX1-1 and BRX1-2 are redundant (Weis et al., 2015b).
Nucleolar Stress Responses in Plant Cells Might Be Mediated by Alternative Routes Rather Than the RP-HDM2-p53 Pathway
We detected the occurrence of nucleolar stress (reduced nucleolar size and growth defect) in the apum24-2 mutant (Figures 9B to 9F) and accompanying accumulation of processing intermediates harboring ITS2 (Figure 9A). Nucleolar stress in plant cells has rarely been investigated to date; there are only a few reports concerning mutations that induce modulations in nucleolar size in plant cells. Qi et al. (2016) reported enlarged nucleolus size in a maize (Zea mays) mutant possessing a mutation in an AAA-ATPase gene involved in the export of the 60S ribosome from the nucleus to the cytoplasm as nucleolar stress. Mutations in two genes, DOMINO and AtREN1, were also reported to cause an increase in nucleolar size (Lahmy et al., 2004; Reňák et al., 2014). However, the physiological relevance of changes in nucleolar size to any internal or external signal responses in plant cells remains unknown. On the other hand, the nucleolus is a key stress-responsive organelle in yeast and animal cells, and the importance of nucleolar stress has already been clarified in animal cells. Nucleolar stress is associated with a variety of growth regulation-associated phenomena, including cell growth/proliferation defects, cell cycle arrest, senescence, and apoptosis in animals (Holmberg Olausson et al., 2012; Zhou et al., 2015). Hence, nucleolar stress likely plays critical roles in plants, even in controlling plant growth. This study showed that reductions in nucleolar size and attenuation of sugar-dependent growth promotion occurred simultaneously in the apum24-2 mutant. The finding that sugar-dependent nucleolar stress occurs in the apum24-2 mutant lays the foundation for exploring new roles of the nucleolus in plant cells.
In human cells, nucleolar stress is mainly transmitted through the RP-HDM2 (human homolog of the MDM2)-p53 pathway. In this pathway, some RPs that accumulate due to dysfunctional ribosome biogenesis are translocated from the nucleoli to the nucleoplasm and subsequently bind to the p53-targeted ubiquitin ligase HDM2 to increase the stability of tumor suppressor protein p53, which regulates cell proliferation arrest, apoptosis, and so on (Zhou et al., 2015). However, plants, like yeast, lack HDM2 and p53 homologs (Rutkowski et al., 2010). Therefore, mechanisms other than the RP-HDM2-p53 pathway are required for the nucleolar stress response in plant cells. Although the RP-HDM2-p53 pathway is a major pathway for inducing the nucleolar stress response in human cells, alternative routes for nucleolar stress responses, including the RPL3-dependent pathway and the RPL11-dependent pathway, have been discovered in humans (Esposito et al., 2014; Challagundla et al., 2011). Therefore, the p53-independent pathways might mediate and induce nucleolar stress responses in plant cells.
Nucleolar Stress Occurs in a Sugar-Dependent Manner in the apum24-2 Mutant
The overaccumulation of processing intermediates containing ITS2 and the occurrence of nucleolar stress in the apum24-2 mutant are sugar-dependent (Figure 9). Since ribosome biogenesis requires a massive amount of energy, and rRNA synthesis is the rate-limiting step of ribosome biogenesis, the sugar-dependent occurrence of nucleolar stress in the apum24-2 mutant might be due to the breakdown of the coordination of ribosome biogenesis-related gene expression with cellular energy status. Therefore, our findings reveal new physiological and molecular connections between nucleolar stress and sugar responses in plants. Although we observed the sugar-dependent occurrence of nucleolar stress in apum24-2 seedlings grown with exogenously supplied sugar (Figure 9), we also found that nucleolar stress in apum24-2 grown in soil under continuous illumination disappeared after 2 d of dark treatment, which promotes the degradation of starch, the storage form of glucose (Supplemental Figures 10B and 10C). We therefore propose that sugar-dependent nucleolar stress is induced and abolished by fluctuations in sugar concentrations in the physiological range.
The transcription of rDNA and ribosome-related genes is promoted by sugar treatment in yeast, mammalian, and plant cells (Powers and Walter, 1999; Iadevaia et al., 2014; Kojima et al., 2007; Ishida et al., 2016). Furthermore, nutrient-dependent rRNA transcription is regulated by various mechanisms, such as chromatin modification at the rDNA locus by histone methylation and the regulation of RNA polymerase I activity by the TOR pathway in mammals (Salifou et al., 2016; Mayer et al., 2004). However, little was known about how the modulation of rRNA processing regulates energy status-responsive ribosome biogenesis. The next challenge is to uncover the effects of nutrients on pre-rRNA processing, ribosome biogenesis, nucleolar stress, and stress-induced responses.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild type, as all T-DNA insertion lines and mutants used in this study were in the Col-0 background. Seeds of apum24-1 (GABI_461E08) and brx1-2 (GABI_771C02) (Weis et al., 2015b) and seeds of apum24-2 (SALK_033623) and brx1-1 (SALK_004020) (Weis et al., 2015b) were provided by GABI-Kat (Rosso et al., 2003) and the Arabidopsis Biological Resource Center (Alonso et al., 2003), respectively. The seeds were sterilized and sown on agar plates of medium containing half-strength Murashige and Skoog salts (1/2 MS) supplemented with various types and concentrations of sugar depending on the experiment. After a 2-d cold treatment, the seeds were germinated and seedlings were grown at 23°C under continuous light (∼70 μmol m−2 s−1; white fluorescence tube lamps; FL40S EX-N TT; Mitsubishi Electric). Healthy seeds that were harvested at the same time from plants that had been grown together in the same growth chamber under identical conditions were used in all the physiological experiments.
Generation of Transgenic Arabidopsis Plants
To produce transgenic plants complemented for the apum24-1 null and apum24-2 knockdown mutations, a genomic DNA fragment for the wild-type APUM24 locus was obtained by PCR using wild-type genomic DNA and four sets of PCR primers (Supplemental Table 2). The four DNA fragments obtained were cloned into pCB302 (Xiang et al., 1999) to reconstruct the wild-type APUM24 locus in a binary vector for complementation tests. The resulting binary vector contained the region from ∼1.4 kb upstream of the translation start codon to ∼1 kb downstream of the stop codon and an insertion of the sequence for three copies of FLAG tag in front of the stop codon. After verification by DNA sequencing, the heterozygote for the apum24-1 allele and the homozygote for the apum24-2 mutant allele were transformed with the binary vector using the floral dip method (Clough and Bent, 1998) with Agrobacterium tumefaciens strain GV3101 (pMP90). To produce binary vectors for the generation of the APUM24-targeted amiRNA lines, DNA fragments spanning the amiRNA target sites (“581 to 601” for the apum24i-1 line and “844 to 864” for the apum24i-2 line; positions are as shown in the APUM24 cDNA sequence) were obtained by performing serial PCRs on wild-type cDNA using two sets of PCR primers listed in Supplemental Table 2. PCRs were performed using the distributed protocol (http://wmd3.weigelworld.org/). The DNA fragments together with the RPS5 promoter fragment, which was also obtained by PCR using specific primers (Supplemental Table 2), were inserted between PstI and EcoRI sites of pCB302 (Xiang et al., 1999). After verification by DNA sequencing, the wild-type plants were transformed with the resultant binary vectors using the floral dip method. Transgenic lines with a T-DNA insertion at a single locus were selected, and T3 progenies homozygous for the inserted gene were selected and used for all experiments.
Proteomic Identification of Proteins in the APUM24-Containing Complex
To prepare the binary vector used to express MYC tag-fused APUM24 in Arabidopsis T87 cells, cDNA for the APUM24 coding region was obtained by RT-PCR using primers APUM24 F’1 and R’1 (Supplemental Table 2), and the EIN3 cDNA in the pHBTpro:EIN3-MYC expression vector (Yanagisawa et al., 2003) was replaced with the APUM24 cDNA. The resulting plasmid, pHBTpro:APUM24-MYC, was used to transform T87 cell suspension cells derived from Arabidopsis ecotype Columbia (Axelos et al., 1992) according to the method of Ogawa et al. (2008). Transformants were selected on JPL medium plates containing 3 g/L gellan gum, 500 mg/L carbenicillin, and 20 mg/L hygromycin and maintained on JPL medium plates without antibiotics.
Suspension cultures of T87 wild-type cells and T87 cells expressing APUM24-MYC were grown in liquid JPL medium. The harvested cells (∼4 g) were frozen in liquid nitrogen and homogenized with a multi-beads shocker (Yasui Kikai) in 10 mL of extraction buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 10% glycerol, and Complete Protease Inhibitor Cocktail [Roche]). The cell lysates were incubated with anti-MYC antibody-cross-linked Dynabeads (Thermo Fisher Scientific), and proteins interacting with the Dynabeads were eluted as described previously (Ishida et al., 2016). NanoLC-ESI-MS/MS analysis of recovered proteins was performed on an electrospray ionization ion trap MS system (LC/MSD Trap XCT Ultra; Agilent Technologies) as described previously (Aki et al., 2008; Aki and Yanagisawa, 2009; Liu et al., 2017). Proteins not detected in T87 wild-type cell lysates but detected in cell lysates from T87 cells expressing APUM24-MYC were selected as candidate interaction partners of APUM24. Because the MS spectral intensities of the respective peptides were proportional to their amounts and the lower limit of intensity for detection was ∼1 × 105 in our MS spectral intensity-based comparative analysis (Hamamoto et al., 2012), the selected proteins were estimated to be more than 100-fold more abundant in the immunoprecipitates obtained from cell lysates containing APUM24-MYC than in the immunoprecipitates obtained from T87 wild-type cell lysates.
Subcellular Localization Analysis via GFP and Split-GFP-Based BiFC Assays
To generate plasmids for the subcellular localization assay and BiFC analysis, cDNAs for the coding regions of APUM24, BRX1-1, BRX1-2, AtLAS1, and AtGRC3 were obtained by RT-PCR of RNA from wild-type Arabidopsis using the primers listed in Supplemental Table 3. The SWA1 and NuGWD1 cDNAs have been described previously (Ishida et al., 2016). The resulting products were cloned into the pENTR/D-TOPO vector (Thermo Fisher Scientific). Then, APUM24, BRX1-1, BRX1-2, and AtLAS1 cDNAs were introduced into pGWB5 to produce GFP C-terminal fusion proteins, whereas AtGRC3 cDNA was introduced into pGWB6 to produce the GFP N-terminal fusion protein. For the BiFC assay, APUM24, SWA1, and AtLAS1 cDNAs were also introduced into pB4GWnG to produce nGFP C-terminal fusion proteins, while AtGRC3 cDNAs were introduced into pB4nGGW to produce nGFP N-terminal fusion proteins. BRX1-1, BRX1-2, NuGWD1, and AtLAS1 cDNAs were introduced into pB4GWcG, while AtGRC3 cDNA was introduced into pB4cGGW to produce cGFP C-terminal fusion or cGFP N-terminal fusion proteins, respectively. Plasmids pGWB5, pGWB6, pB4GWnG, pB4nGGW, pB4GWcG, and pB4cGGW were described by Tanaka et al. (2012), and LR Clonase II (Thermo Fisher Scientific) was used to produce the constructs. All PCR products and inserts were verified by DNA sequencing. The SWA1 and NuGWD1 cDNAs were introduced into pB4GWcG and pB4GWnG, as described previously (Ishida et al., 2016).
The subcellular localization assay and split-GFP-based BiFC assay were performed by coinfiltrating Nicotiana benthamiana leaves (Kapila et al., 1997; D’Aoust et al., 2009) with derivatives of pGWB5, pGWB6, pB4GWnG, pB4nGGW, pB4GWcG, and pB4cGGW as described previously (Maekawa et al., 2014). The FIB1-mCherry expression vector was cotransformed with the derivatives to identify transformed cells and to clarify the location of the nucleolus (Maekawa et al., 2014). Two days after infection, GFP fluorescence was observed under a fluorescence microscope (BX51; Olympus) equipped with a cooled color digital camera (DP80; Olympus).
Microscopy of Leaf Shape
First or second leaves were fixed in 100% ethanol, dehydrated through a graded ethanol series, and mounted in clearing solution (chloral hydrate, glycerol, and water at a ratio of 8 g:1 mL:2 mL). Images were obtained under a stereomicroscope (MZ16F; Leica Microsystems) equipped with a cooled color digital camera (DXM1200C; Nikon Instruments).
RT-qPCR Analysis
To quantify the expression levels of sugar-responsive genes, surface-sterilized seeds were soaked in 50 mL of one-tenth-strength MS liquid medium supplemented with 1 g L−1 sucrose. After 2 d of cold treatment, the seeds were grown at 23°C under continuous light conditions with gentle shaking for 1 week. The cultured young seedlings were washed well with one-tenth-strength MS liquid medium and cultured in 50 mL of one-tenth-strength MS liquid medium in the dark for 1 d with gentle shaking. Various sugars (mannitol, fructose, glucose, and sucrose) were added to the medium at a final concentration of 50 mM, and the seedlings were harvested 3 h later. To monitor the pre-rRNA processing pattern, seedlings were grown on 1/2 MS medium plates with or without glucose or mannitol under diurnal light conditions (16 h/8 h).
RNA was prepared from seedlings using an ISOSPIN Plant RNA kit (Nippon Gene) with a TURBO DNase kit (Thermo Fisher Scientific), and reverse transcription was performed with SuperScript II reverse transcriptase (Thermo Fisher Scientific) with random primers. PCR was performed with a StepOne Plus Real-Time PCR System (Thermo Fisher Scientific) using a KAPA SYBR Fast Quantitative PCR kit (KAPA Biosystems). The primer sequences and the efficiency of each PCR are listed in Supplemental Table 3. Relative gene expression levels were calculated using the ΔΔCT method (Livak and Schmittgen, 2001) and normalized relative to the expression levels of UBQ10.
Measurement of Anthocyanin Contents
Anthocyanin contents were determined as described by Mehrtens et al. (2005) with minor modifications. In brief, ∼10 mg of whole plant tissue was submerged in 600 μL of acidic methanol (1% HCl, w/v) and incubated overnight at 4°C with gentle shaking. Anthocyanin was extracted with 400 μL of distilled water and 400 μL of chloroform. The absorption of the extracts at 530 and 657 nm was measured, and anthocyanin levels were calculated using the equation A530 – (A675 × 0.25).
RIP Assay
The RIP assay was performed basically as described by Saleh et al. (2008) and Terzi and Simpson (2009). In brief, ∼200 seedlings (16 d old) were submerged and permeated with cross-linking buffer (10 mM Tris-HCl, pH 8.0, 0.4 M sucrose, 1 mM EDTA, 1% formaldehyde, 1 mM PMSF, and 1× Complete protease inhibitor) via vacuum infiltration for 15 min; the reaction was stopped by adding glycine (final concentration of 200 mM). The tissues were ground to a fine powder in liquid nitrogen and suspended in 7.5 mL of extraction buffer (10 mM Tris-HCl, pH 8.0, 0.4 M sucrose, 5 mM DTT, 1 mM PMSF, 1× Complete protease inhibitor, and 200 units of SUPERase In RNase Inhibitor [Thermo Fisher Scientific]). The slurry was filtered through two layers of Miracloth (Millipore), and MgCl2 and Triton X-100 were added to the filtrates to a final concentration of 10 mM and 0.2%, respectively. After 10 min incubation, nuclei were recovered by centrifugation and suspended in 1 mL of nuclear lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.3% SDS, 1 mM PMSF, 1× Complete protease inhibitor, and 40 units of SUPERase In RNase Inhibitor). After sonication to shear RNA and the removal of insoluble materials by centrifugation, the nuclear lysate was mixed with 30 μL of protein A agarose beads (Roche) and incubated for 1 h at 4°C to remove proteins that nonspecifically interacted with the protein A agarose beads. An aliquot (130 μL) of the nuclear lysate was preserved as an input sample, and the remainder was mixed with 30 μL of protein A agarose beads conjugated with anti-DYKDDDDK antibody (Wako) and incubated for 16 h. The immune complexes were eluted with 130 μL of elution buffer (1% SDS, 0.1 M NaHCO3, and 5 units of SUPERase In RNase Inhibitor). For proteolysis and reversal of cross-linking, 5 μL of 0.5 M EDTA (pH 8.0), 10 μL of 1 M Tris-HCl (pH 6.5), 1.3 μL of 20 mg/mL proteinase K, and 10 μL of 5 M NaCl were added to the input and eluted samples, which were then incubated at 42°C for 1 h and 60°C for 1 h. RNA was recovered from the samples with ISOGEN solution and a TURBO DNase kit, and reverse transcription was performed with PrimeScript RT Master Mix (Takara). PCR was performed as described above (for RT-qPCR) using rRNA-related PCR primers (Supplemental Table 3). The fold enrichment was calculated as the ratio of the RNA level from the eluate to that from the input sample.
RNA Gel Blot Analysis
Total RNA was isolated from 7-d-old seedlings grown on 1/2 MS solid medium with 50 mM glucose using an ISOSPIN Plant RNA kit (Nippon Gene) and a TURBO DNase kit (Thermo Fisher Scientific). Total RNA (4 μg) was separated on a 1% agarose gel containing formaldehyde and transferred to a positively charged nylon membrane (Hybond N+) (GE Healthcare) by capillary transfer. Probe DNAs were prepared by PCR with appropriate primers (Supplemental Table 2). DIG-labeling of probe DNAs, hybridization, and detection were performed using a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) following the manufacturer’s instructions.
cRT-PCR Analysis
cRT-PCR analysis was performed according to the method of Abbasi et al. (2010). Total RNA (1.5 μg) was prepared from 7-d-old seedlings grown on 1/2 MS solid medium with 50 mM glucose using an ISOSPIN Plant RNA kit and a TURBO DNase kit and circularized with T4 RNA ligase (Takara Bio). To detect RNA containing the 25S rRNA sequence, the first-strand cDNA was synthesized with circularized RNA and the 25S-cRT primer, followed by PCR using 25S-L and 25S-R PCR primers. To detect RNA containing the 18S rRNA sequence, the first-strand cDNA was synthesized with the 18S-cRT primer, followed by PCR using 18S-L and 18S-R primers. After a 35-cycle amplification, the cRT-PCR products were analyzed by 2% agarose gel electrophoresis and the DNA bands were excised. The DNA fragments recovered were cloned into the pENTR D-TOPO vector using a TOPO TA cloning kit (Life technologies). Three independent clones per cRT-PCR product were sequenced using M13F and M13R primers. Primer sequences are listed in Supplemental Table 2.
Measurement of Nucleolar Size
To measure nucleolar size, expression vectors for GFP fused to the C terminus of FIB1 (FIB1-GFP) or SWA1 (SWA1-GFP) were generated by LR reactions using the entry vector pUGW5 (Tanaka et al., 2012) and FIB1 and SWA1 cDNAs (Ishida et al. 2016) with LR Clonase II. The resulting vectors were verified by DNA sequencing and used for transient expression of FIB1-GFP and SWA1-GFP in Arabidopsis mesophyll protoplasts. The protoplasts were isolated from young seedlings grown on 1/2 MS medium plates with or without 50 mM glucose or plants grown in soil for 4 weeks with an additional 2 d in the dark or under continuous light. Transient expression in Arabidopsis mesophyll protoplasts was performed as described by Yoo et al. (2007) and Wu et al. (2009) with minor modifications. After transfection, the protoplasts were incubated for 16 h at room temperature in the dark. Images of GFP fluorescence were taken under a fluorescence microscope (BX51) equipped with a cooled color digital camera (DP80) and utilized for nucleolus areas calculations with Image J (Schneider et al., 2012).
Histochemical Detection of Starch
Detached leaves were depigmented with 5 mL of 100% ethanol in six-well plates for ∼16 h, followed by three washes with 5 mL of water. The leaves were then stained with 2 mL of Lugol’s iodine solution (Sigma-Aldrich) for 10 min, followed by two washes with 5 mL of water to remove the remaining solution.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: APUM24 (At3g16810), BRX1-1 (At3g15460), BRX1-2 (At1g52930), FIB1 (At5g52470), RPS8A (At5g20290), RPL4A (At3g09630), AtLAS1 (At5g12220), AtGRC3 (At5g11010), SWA1 (At2g47990), and NuGWD1 (At5g11240).
Supplemental Data
Supplemental Figure 1. Two Independent APUM24-Targeted amiRNA Lines Display Phenotypes Resembling Those of the apum24-2 Mutant.
Supplemental Figure 2. Coexpression Network Including APUM24, BRX1-1, and BRX1-2.
Supplemental Figure 3. Negative Controls for BiFC Analyses.
Supplemental Figure 4. The brx1-1 as2 Double Mutant Displays Pointed True Leaves with Serrated Edges.
Supplemental Figure 5. Increased Antibiotic Resistances in apum24-2 and brx1-2.
Supplemental Figure 6. Specific Immunoprecipitation of APUM24-FLAG Protein for the RIP Assay.
Supplemental Figure 7. Alignment of Amino Acid Sequences from AtLAS1, HsLAS1, and ScLas1.
Supplemental Figure 8. AtGRC3, HsGRC3, and ScGrc3 Amino Acid Sequence Alignments.
Supplemental Figure 9. Coexpression Network Including APUM24, AtLAS1, and AtGRC3.
Supplemental Figure 10. Sugar-Dependent and Light Treatment-Dependent Nucleolar Stress in the apum24-2 Mutant.
Supplemental Table 1. Results of BLAST Searches to Identify Arabidopsis Homologs of Components of the Yeast C2 Endonuclease Complex.
Supplemental Table 2. List of Primers Used for Vector Construction, Genotyping, RT-PCR, RNA Gel Blot, and cRT-PCR.
Supplemental Table 3. List of Primers Used for RT-qPCR.
Supplemental Table 4. ANOVA Results.
Acknowledgments
We thank Junji Yamaguchi (Hokkaido University, Japan), Shoji Mano (National Institute of Basic Biology, Japan), Tsuyoshi Nakagawa (Shimane University, Japan), the Arabidopsis Biological Resource Center, GABI-Kat, and the RIKEN BioResource Center for kindly providing the entry vector for FIB1 and the 35Spro:FIB1-mCherry vector, Gateway vectors for GFP-fused protein expression and BiFC assays, seeds of the SALK T-DNA collection lines, seeds of the GABI-Kat collection lines, and T87 cells, respectively. This work was supported in part by JST CREST Grant JPMJCR15O5 and JSPS KAKENHI Grants JP25252014 and JP26221103 to S.Y. and by JSPS KAKENHI Grant JP15J08368 to S.M.
AUTHOR CONTRIBUTIONS
S.M., T.I., and S.Y. designed the research. S.M. and T.I. performed the experiments and analyzed the data. S.M. and S.Y. wrote the article.
References
- Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64: 960–976. [DOI] [PubMed] [Google Scholar]
- Aki T., Shigyo M., Nakano R., Yoneyama T., Yanagisawa S. (2008). Nano scale proteomics revealed the presence of regulatory proteins including three FT-Like proteins in phloem and xylem saps from rice. Plant Cell Physiol. 49: 767–790. [DOI] [PubMed] [Google Scholar]
- Aki T., Yanagisawa S. (2009). Application of rice nuclear proteome analysis to the identification of evolutionarily conserved and glucose-responsive nuclear proteins. J. Proteome Res. 8: 3912–3924. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Makarova K.S., Burroughs A.M., Koonin E.V., Aravind L (2013). Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct. 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armistead J., Triggs-Raine B. (2014). Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett. 588: 1491–1500. [DOI] [PubMed] [Google Scholar]
- Axelos M., Curic C., Mazzolini L., Bardet C., Lescure B. (1992). A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol. Biochem. 30: 123–128. [Google Scholar]
- Barneche F., Steinmetz F., Echeverría M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275: 27212–27220. [DOI] [PubMed] [Google Scholar]
- Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. (2010). The nucleolus under stress. Mol. Cell 40: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A.L., David R., Searle I.R. (2015). Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 15: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E. (2009). A role for the ribosome in development. Trends Plant Sci. 14: 512–519. [DOI] [PubMed] [Google Scholar]
- Chakraborty A., Uechi T., Kenmochi N. (2011). Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip. Rev. RNA 2: 507–522. [DOI] [PubMed] [Google Scholar]
- Challagundla K.B., Sun X.X., Zhang X., DeVine T., Zhang Q., Sears R.C., Dai M.S. (2011). Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol. Cell. Biol. 31: 4007–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.Y., Fan C.C., Chu P.C., Hong B.E., Lee H.J., Chang M.S. (2011). hPuf-A/KIAA0020 modulates PARP-1 cleavage upon genotoxic stress. Cancer Res. 71: 1126–1134. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- D’Aoust M.A., Lavoie P.O., Belles-Isles J., Bechtold N., Martel M., Vézina L.P. (2009). Transient expression of antibodies in plants using syringe agroinfiltration. Methods Mol. Biol. 483: 41–50. [DOI] [PubMed] [Google Scholar]
- Esposito D., Crescenzi E., Sagar V., Loreni F., Russo A., Russo G. (2014). Human rpL3 plays a crucial role in cell response to nucleolar stress induced by 5-FU and L-OHP. Oncotarget 5: 11737–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C.C., Lee L.Y., Yu M.Y., Tzen C.Y., Chou C., Chang M.S. (2013). Upregulated hPuf-A promotes breast cancer tumorigenesis. Tumour Biol. 34: 2557–2564. [DOI] [PubMed] [Google Scholar]
- Francischini C.W., Quaggio R.B. (2009). Molecular characterization of Arabidopsis thaliana PUF proteins--binding specificity and target candidates. FEBS J. 276: 5456–5470. [DOI] [PubMed] [Google Scholar]
- Gasse L., Flemming D., Hurt E. (2015). Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell 60: 808–815. [DOI] [PubMed] [Google Scholar]
- Gu W., Deng Y., Zenklusen D., Singer R.H. (2004). A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 18: 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto K., Aki T., Shigyo M., Sato S., Ishida T., Yano K., Yoneyama T., Yanagisawa S. (2012). Proteomic characterization of the greening process in rice seedlings using the MS spectral intensity-based label free method. J. Proteome Res. 11: 331–347. [DOI] [PubMed] [Google Scholar]
- Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg Olausson K., Nistér M., Lindström M.S. (2012). p53-dependent and -independent nucleolar stress responses. Cells 1: 774–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Van Lijsebettens M., Candela H., Micol J.L., Tsukaya H. (2012). Ribosomes and translation in plant developmental control. Plant Sci. 191-192: 24–34. [DOI] [PubMed] [Google Scholar]
- Huang T., Kerstetter R.A., Irish V.F. (2014). APUM23, a PUF family protein, functions in leaf development and organ polarity in Arabidopsis. J. Exp. Bot. 65: 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.U., Kim M.J., Paek K.H. (2013). Arabidopsis Pumilio protein APUM5 suppresses Cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. USA 110: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SU, Paek KH (2014). APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 14: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V., Liu R., Proud C.G. (2014). mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 36: 113–120. [DOI] [PubMed] [Google Scholar]
- Ishida T., Maekawa S., Yanagisawa S. (2016). The pre-rRNA processing complex in Arabidopsis includes two WD40-domain-containing proteins encoded by glucose-inducible genes and plant-specific proteins. Mol. Plant 9: 312–315. [DOI] [PubMed] [Google Scholar]
- James A., Wang Y., Raje H., Rosby R., DiMario P. (2014). Nucleolar stress with and without p53. Nucleus 5: 402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J.L., Guan K.L. (2013). Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 38: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila J., DeRycke R., VanMontagu M., Angenon G. (1997). An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122: 101–108. [Google Scholar]
- Kaser A., Bogengruber E., Hallegger M., Doppler E., Lepperdinger G., Jantsch M., Breitenbach M., Kreil G. (2001). Brix from Xenopus laevis and brx1p from yeast define a new family of proteins involved in the biogenesis of large ribosomal subunits. Biol. Chem. 382: 1637–1647. [DOI] [PubMed] [Google Scholar]
- Kojima H., Suzuki T., Kato T., Enomoto K., Sato S., Kato T., Tabata S., Sáez-Vasquez J., Echeverría M., Nakagawa T., Ishiguro S., Nakamura K. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Kressler D., Hurt E., Bassler J. (2010). Driving ribosome assembly. Biochim. Biophys. Acta 1803: 673–683. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L. (2010). A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem. Sci. 35: 267–277. [DOI] [PubMed] [Google Scholar]
- Lahmy S., Guilleminot J., Cheng C.M., Bechtold N., Albert S., Pelletier G., Delseny M., Devic M. (2004). DOMINO1, a member of a small plant-specific gene family, encodes a protein essential for nuclear and nucleolar functions. Plant J. 39: 809–820. [DOI] [PubMed] [Google Scholar]
- Lee I., Li Z., Marcotte E.M. (2007). An improved, bias-reduced probabilistic functional gene network of baker’s yeast, Saccharomyces cerevisiae. PLoS One 2: e988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H., Shore D. (2009). Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21: 855–863. [DOI] [PubMed] [Google Scholar]
- Li Z., Lee I., Moradi E., Hung N.J., Johnson A.W., Marcotte E.M. (2009). Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 7: e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.-H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Loewith R., Hall M.N. (2011). Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida C., Nakagawa A., Kojima S., Takahashi H., Machida Y. (2015). The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 4: 655–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Inada N., Yasuda S., Fukao Y., Fujiwara M., Sato T., Yamaguchi J. (2014). The carbon/nitrogen regulator ARABIDOPSIS TOXICOS EN LEVADURA31 controls papilla formation in response to powdery mildew fungi penetration by interacting with SYNTAXIN OF PLANTS121 in Arabidopsis. Plant Physiol. 164: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T., Oswald O., Graham I.A. (2002). Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128: 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y., et al. (2016). A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol. Open 5: 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Zhao J., Yuan X., Grummt I. (2004). mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P., Weisshaar B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138: 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A., et al. (2008). Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133: 627–639. [DOI] [PubMed] [Google Scholar]
- Nicolas E., Parisot P., Pinto-Monteiro C., de Walque R., De Vleeschouwer C., Lafontaine D.L. (2016). Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 7: 11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35: D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y., Dansako T., Yano K., Sakurai N., Suzuki H., Aoki K., Noji M., Saito K., Shibata D. (2008). Efficient and high-throughput vector construction and Agrobacterium-mediated transformation of Arabidopsis thaliana suspension-cultured cells for functional genomics. Plant Cell Physiol. 49: 242–250. [DOI] [PubMed] [Google Scholar]
- Powers T., Walter P. (1999). Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J., Laxmi A., St Martin S.K., Jang J.C. (2004). Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Zhu J., Wu Q., Wang Q., Li X., Yao D., Jin Y., Wang G., Wang G., Song R. (2016). Maize reas1 mutant stimulates ribosome use efficiency and triggers distinct transcriptional and translational responses. Plant Physiol. 170: 971–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reňák D., Gibalová A., Solcová K., Honys D. (2014). A new link between stress response and nucleolar function during pollen development in Arabidopsis mediated by AtREN1 protein. Plant Cell Environ. 37: 670–683. [DOI] [PubMed] [Google Scholar]
- Rosado A., Sohn E.J., Drakakaki G., Pan S., Swidergal A., Xiong Y., Kang B.H., Bressan R.A., Raikhel N.V. (2010). Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell 22: 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259. [DOI] [PubMed] [Google Scholar]
- Rutkowski R., Hofmann K., Gartner A. (2010). Phylogeny and function of the invertebrate p53 superfamily. Cold Spring Harb. Perspect. Biol. 2: a001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Salifou K., Ray S., Verrier L., Aguirrebengoa M., Trouche D., Panov K.I., Vandromme M. (2016). The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nat. Commun. 7: 10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D.Q., Liu J., Xiang Y.H., Ye D., Sundaresan V., Yang W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov D.S., Jurecic R. (2003). The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 55: 359–366. [DOI] [PubMed] [Google Scholar]
- Tam P.P., Barrette-Ng I.H., Simon D.M., Tam M.W., Ang A.L., Muench D.G. (2010). The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Kimura T., Hikino K., Shino Goto S., Nishimura M., Mano S. Nakagawa T. (2012). Gateway vectors for plant genetic engineering: Overview of plant vectors, application for bimolecular fluorescence complementation (BiFC) and multigene construction. In Genetic Engineering: Basics, New Applications and Responsibilities, Barrera-Saldana H.A., ed (Croatia: InTech), pp. 35–58. [Google Scholar]
- Terzi L.C., Simpson G.G. (2009). Arabidopsis RNA immunoprecipitation. Plant J. 59: 163–168. [DOI] [PubMed] [Google Scholar]
- Tschochner H., Hurt E. (2003). Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13: 255–263. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Kovacevic J., Missbach S., Schleiff E. (2015a). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20: 729–740. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Palm D., Missbach S., Bohnsack M.T., Schleiff E. (2015b). atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Bernstein D.S., Kimble J., Parker R. (2002). A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 18: 150–157. [DOI] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Han P., Lutziger I., Wang K., Oliver D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Nakabayashi K., Ding J., He F., Bentsink L., Soppe W.J. (2014). Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26: 4362–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Song B., Née G., Kramer K., Finkemeier I., Soppe W.J. (2016). Sequence polymorphisms at the REDUCED DORMANCY5 pseudophosphatase underlie natural variation in Arabidopsis dormancy. Plant Physiol. 171: 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Zhao H., Li Q.Q. (2008). Arabidopsis CLP1-SIMILAR PROTEIN3, an ortholog of human polyadenylation factor CLP1, functions in gametophyte, embryo, and postembryonic development. Plant Physiol. 148: 2059–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Sheen J. (2014). The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 164: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.T., Ting Y.H., Liang K.J., Lo K.Y. (2016). The roles of Puf6 and Loc1 in 60S biogenesis are interdependent, and both are required for efficient accommodation of Rpl43. J. Biol. Chem. 291: 19312–19323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S., Yoo S.D., Sheen J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang C., Muench D.G. (2015). A nucleolar PUF RNA-binding protein with specificity for a unique RNA sequence. J. Biol. Chem. 290: 30108–30118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liao W.J., Liao J.M., Liao P., Lu H. (2015). Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]