MUT9p-LIKE PROTEIN1 (MLK1) and MLK2 mediate the interaction between gibberellins and the circadian clock, thereby regulating plant growth in Arabidopsis.
Abstract
Gibberellins (GAs) modulate diverse developmental processes throughout the plant life cycle. However, the interaction between GAs and the circadian rhythm remains unclear. Here, we report that MUT9p-LIKE KINASE1 (MLK1) and MLK2 mediate the interaction between GAs and the circadian clock to regulate hypocotyl elongation in Arabidopsis thaliana. DELLA proteins function as master growth repressors that integrate phytohormone signaling and environmental pathways in plant development. MLK1 and MLK2 interact with the DELLA protein REPRESSOR OF ga1-3 (RGA). Loss of MLK1 and MLK2 function results in plants with short hypocotyls and hyposensitivity to GAs. MLK1/2 and RGA directly interact with CIRCADIAN CLOCK ASSOCIATED1 (CCA1), which targets the promoter of DWARF4 (DWF4) to regulate its roles in cell expansion. MLK1/2 antagonize the ability of RGA to bind CCA1, and these factors coordinately regulate the expression of DWF4. RGA suppressed the ability of CCA1 to activate expression from the DWF4 promoter, but MLK1/2 reversed this suppression. Genetically, MLK1/2 act in the same pathway as RGA and CCA1 in hypocotyl elongation. Together, our results provide insight into the mechanism by which MLK1 and MLK2 antagonize the function of RGA in hypocotyl elongation and suggest that MLK1/2 coordinately mediate the regulation of plant development by GAs and the circadian rhythm in Arabidopsis.
INTRODUCTION
In seedlings that germinate underground, hypocotyl elongation helps the shoot to reach the surface of the soil, pushing the cotyledons into the light and enabling the switch to autotrophy. Hypocotyl elongation is controlled by endogenous regulators, such as phytohormones and the circadian clock, as well as environmental stimuli such as light signaling, touch, and temperature (Saibo et al., 2003). Brassinosteroids, auxin, and gibberellins (GAs) promote hypocotyl growth, whereas cytokinins and abscisic acid inhibit hypocotyl growth (Clouse, 1996; Gray et al., 1998). GAs control many aspects of plant development, including seed germination, leaf expansion, stem elongation, flowering, and seed development (Sun and Gubler, 2004; Davière and Achard, 2013). GAs promote hypocotyl growth via cell elongation and are strictly required for hypocotyl elongation in dark-grown seedlings (Cowling and Harberd, 1999). However, in Arabidopsis thaliana, brassinosteroids can overcome the lack of GAs and promote elongation in darkness and in the light (Bai et al., 2012; Gallego-Bartolomé et al., 2012). The GA signaling pathway is controlled by the DELLA repressors, which have a characteristic N-terminal DELLA domain. The Arabidopsis genome encodes five DELLA proteins, namely, GA INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3, and the rice (Oryza sativa) genome encodes one DELLA protein, SLENDER RICE1 (SLR1) (Ikeda et al., 2001). All of these DELLA proteins function as negative regulators of GA signaling (Olszewski et al., 2002). RGA and GAI redundantly repress elongation growth (Dill and Sun, 2001), whereas RGL1 and RGL2 primarily function in seed germination and floral development (Lee et al., 2002; Wen and Chang, 2002; Cheng et al., 2004).
In the absence of GA, DELLA proteins interact with transcription factors to inhibit the transcription of GA-responsive genes (Sun and Gubler, 2004; Feng et al., 2008; Sun, 2011). In response to GA, DELLA proteins are inactivated through binding with GIBBERELLIN INSENSITIVE DWARF1 (GID1) (Ariizumi et al., 2008) and are ubiquitinated by the SCFSLY1/GID2 (Skp1-Cullin-F-box protein complex) E3 ligase, followed by degradation by the 26S proteasome system, triggering GA responses (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Gomi et al., 2004; Sun, 2011). Earlier studies have suggested that GA-induced phosphorylation of DELLAs is a prerequisite for their degradation (Sasaki et al., 2003; Fu et al., 2004), but subsequent work demonstrated that phosphorylation of DELLAs does not depend on GA and that GID2 recognition does not require the phosphorylation of SLR1, which is the only DELLA protein in rice (Itoh et al., 2005; Davière et al., 2008). Although the functional importance of DELLA phosphorylation had been unclear (Nelson and Steber, 2016), subsequent studies have shown that the phosphorylation of DELLA proteins is essential for their stability. In rice, the Ser/Thr casein kinase I EARLY FLOWERING1 (EL1) phosphorylates SLR1, and the el1 loss-of-function mutant exhibits enhanced GA-modulated degradation of DELLAs (Dai and Xue, 2010). Analysis of Arabidopsis TOPP4 provided further evidence that phosphorylation positively regulates the repression of GA signaling by DELLAs, whereas dephosphorylation negatively regulates this process (Qin et al., 2014; Nelson and Steber, 2016). Brassinosteroids promote hypocotyl elongation. DWARF4 (DWF4) encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis (Choe et al., 1998). Loss of DWF4 function results in a dwarf phenotype with a defect in cell elongation, and the dwarf phenotype was rescued by exogenous brassinolide application (Azpiroz et al., 1998).
In addition to GAs and brassinosteroids, the circadian clock regulates hypocotyl elongation. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), two homologous MYB-domain transcription factors that partially overlap in function, accumulate at dawn and form a central loop in circadian regulation (Schaffer et al., 1998; Wang and Tobin, 1998; Mizoguchi et al., 2002). The circadian clock regulates the expression of two growth-promoting transcription factor genes involved in cell elongation, PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5, when plants are in darkness. This regulation occurs through a complex comprising the evening-expressed proteins EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRHYTHMO (Nusinow et al., 2011). CCA1 represses ELF3 by associating with its promoter, completing a CCA1-ELF3 negative feedback loop that places ELF3 within the circadian oscillator. ELF3 acts downstream of CCA1, mediating the repression of PIF4 and PIF5 in the control of hypocotyl elongation (Lu et al., 2012). Recent evidence showed that ELF3 inhibits PIF4 activity via a direct interaction and suppresses PIF4 transcriptional activity (Nieto et al., 2015).
DELLAs integrate other phytohormone signaling pathways and environmental pathways to regulate plant development and defense (Harberd et al., 2009; Sun, 2011), including auxin, ethylene, abscisic acid, brassinosteroid, and jasmonate signaling (Weiss and Ori, 2007; Hou et al., 2010; An et al., 2012; Bai et al., 2012), as well as environmental responses to light (de Lucas et al., 2008; Feng et al., 2008), cold (Achard et al., 2008b), and salt (Achard et al., 2008a). In addition, DELLAs interact with a number of transcription factors and directly inactivate these factors (Weiss and Ori, 2007; Feng et al., 2008; Hou et al., 2010; Zhang et al., 2011; Bai et al., 2012). These transcription factors include the bHLH factor PIF4, which promotes cell elongation when plants are in darkness or shade, or at high temperature (Feng et al., 2008), JASMONATE ZIM-DOMAIN1, a key repressor of jasmonate signaling (Hou et al., 2010), and BRASSINAZOLE-RESISTANT1, which controls brassinosteroid-responsive gene expression. DELLAs modulate jasmonate signaling via competitive binding to JAZs (Bai et al., 2012). Despite their overlapping physiological functions and our extensive knowledge of each individual signaling pathway, little is known about how GA and the circadian clock interact at the molecular level.
Casein kinase I, a serine/threonine protein kinase, is a multifunctional protein kinase found in most eukaryotic cells. In mammalian cells, casein kinase I is involved in vesicular trafficking, DNA repair, circadian rhythm, and morphogenesis (McKay et al., 2001). In rice, EL1 encodes a casein kinase I that phosphorylates the rice DELLA protein SLR1 (Dai and Xue, 2010). In the alga Chlamydomonas reinhardtii, MUT9p is related to casein kinase I and phosphorylates H3 at threonine 3 (Casas-Mollano et al., 2008). The Arabidopsis genome encodes four proteins (MLK1, MLK2, MLK3, and MLK4) related to MUT9p (Wang et al., 2015; Huang et al., 2016; Su et al., 2017). MLK1 and MLK2 were first identified as kinases for phosphorylation of H3 at threonine 3 and are associated with the osmotic stress response (Wang et al., 2015), while MLK4 was characterized as a kinase for phosphorylation of H2A at serine 95 and promoted flowering time under long-day conditions (Su et al., 2017). A recent study showed that MLK1 and MLK2 copurified with components of the evening complex of the circadian clock and with phytochrome B (Huang et al., 2016). Loss of MLK1 and MLK2 function results in short hypocotyls and period lengthening of the circadian clock (Huang et al., 2016). Although hypocotyl elongation is controlled by phytohormones and the circadian clock, how phytohormones and the circadian clock interact genetically and in the control of physiological processes of hypocotyl elongation has not been elucidated. Here, we report that MLK1/2 promote hypocotyl elongation by enhancing cell elongation. Our results showed that MLK1/2 act antagonistically to RGA to bind CCA1 and coordinately regulate the expression of DWF4 and hypocotyl elongation in Arabidopsis.
RESULTS
MLK1 and MLK2 Are Required for Hypocotyl Elongation
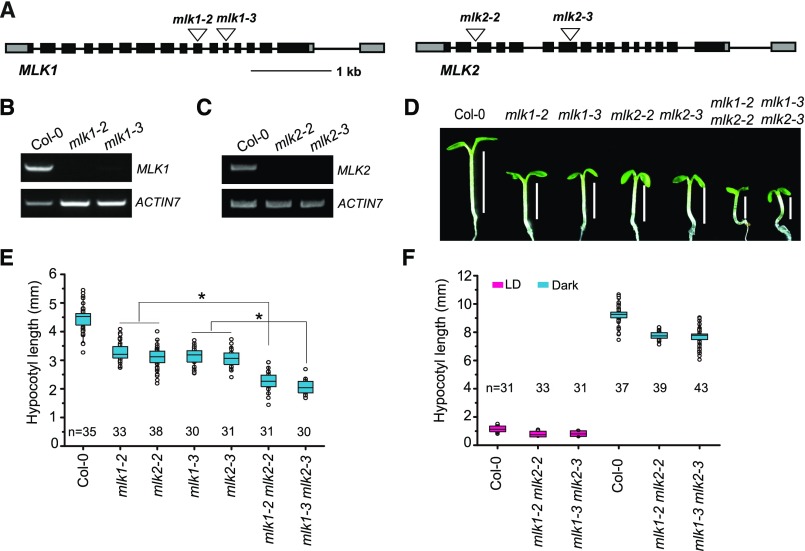
To functionally characterize MLK1 (AT5G18190) and MLK2 (AT3G03940), we identified mlk1 and mlk2 T-DNA insertion mutants. Genotypic analyses revealed the presence of a T-DNA insertion in exon 10 and exon 12 of MLK1 in the mlk1-2 (Salkseq_12450) and mlk1-3 (SALK_039903) mutants and in intron 2 and exon 7 of MLK2 in the mlk2-2 (SALK_149222) and mlk2-3 (SALK_064333) mutants, respectively (Figure 1A). No full-length MLK1 or MLK2 transcripts were detected in the mlk1 or mlk2 mutants, indicating that both mutants are null alleles (Figures 1B and 1C).
Figure 1.
Mutations in MLK1 and MLK2 Result in Short Hypocotyls.
(A) Gene structures of MLK1 and MLK2, including exons (boxes), introns (lines), and T-DNA insertions (triangles).
(B) and (C) RT-PCR analysis of MLK1 and MLK2 expression in mlk1 and mlk2 mutants.
(D) Representative images of Col-0, mlk1, mlk2, and mlk1 mlk2 plants under SD conditions.
(E) Hypocotyls of Col-0, mlk1, mlk2, and mlk1 mlk2 plants under SDs. Values shown are mean number ± sd of hypocotyl length. Asterisks indicate significant difference using Student’s t test (P < 0.05).
(F) Hypocotyls of Col-0, mlk1, mlk2, and mlk1 mlk2 plants under LD conditions and in darkness. The hypocotyl lengths were indicated with lines. Values shown are mean number ± sd of hypocotyl length.
The mlk1 and mlk2 mutants had short hypocotyls under short-day (SD) conditions (Figures 1D and 1E). To evaluate the redundant functions of MLK1 and MLK2, we generated two double mutants by crossing mlk1-2 with mlk2-2 and mlk1-3 with mlk2-3. The mlk1-2 mlk2-2 and mlk1-3 mlk2-3 double mutants had shorter hypocotyls than the mlk1 and mlk2 single mutants. Short hypocotyls were also observed under long-day (LD) conditions and in the dark, suggesting that MLK1 and MLK2 might be involved in the GA pathway (Figure 1F). The redundant functions of MLK1 and MLK2 are supported by the observation that the mlk1 mlk2 double mutants, but not the mlk1 or mlk2 single mutants, exhibited late flowering under LD conditions (Supplemental Figure 1).
MLK1 and MLK2 Are Involved in the GA Pathway
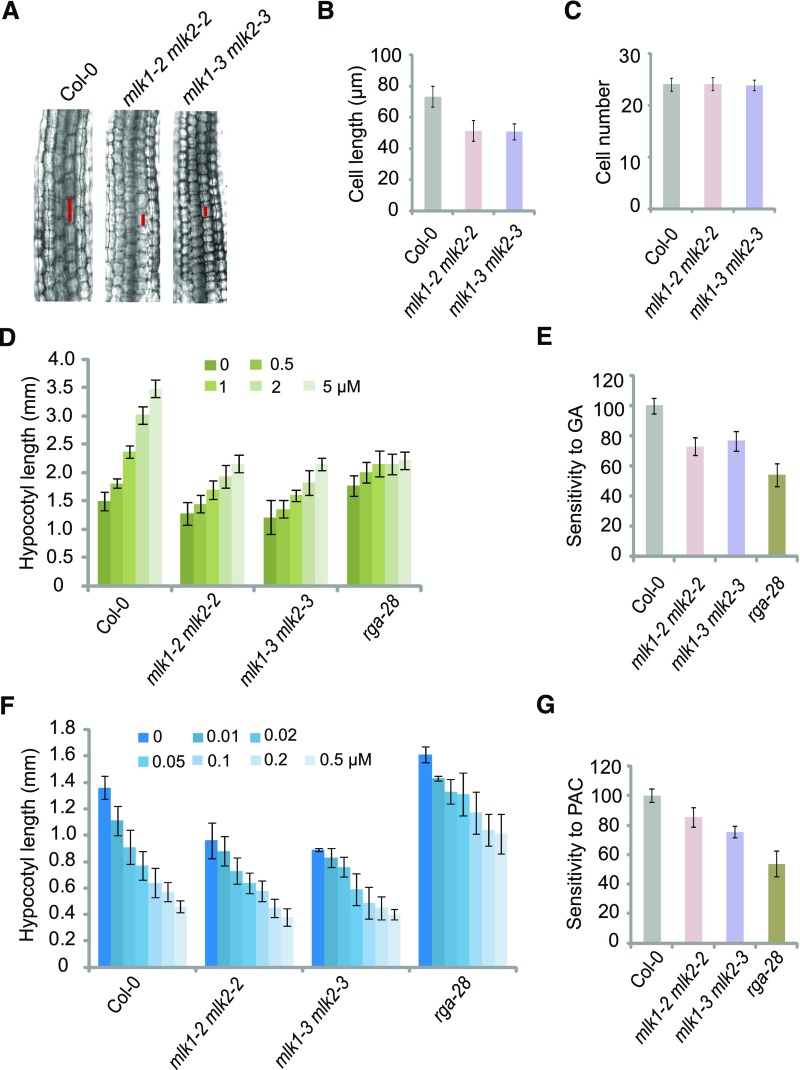
To investigate the roles of MLK1 and MLK2 in the GA pathway, we examined the cell length and cell number in hypocotyls of wild-type and mlk1 mlk2 double mutant plants. The cell length, but not the cell number, was reduced in mlk1 mlk2 seedlings (Figures 2A to 2C). We then investigated the response of the mlk1 mlk2 double mutants to GAs. In the presence of exogenous GAs, the rate of elongation and the final lengths of hypocotyls increased in both wild-type and mlk1 mlk2 plants. However, compared with the wild type, mlk1 mlk2 exhibited a reduced GA response (Figures 2D and 2E), suggesting that MLK1 and MLK2 are involved in GA signal transduction. We treated wild-type and mlk1 mlk2 plants with paclobutrazol (PAC), an inhibitor of GA biosynthesis. The mlk1 mlk2 double mutants were less sensitive to PAC treatment than wild-type seedlings based on hypocotyl elongation (Figures 2F and 2G). Therefore, MLK1 and MLK2 are involved in GA signal transduction.
Figure 2.
The mlk1 mlk2 Double Mutants Are Hyposensitive to GAs.
(A) Cell lengths in hypocotyls of 1-week-old Col-0 and mlk1 mlk2 double mutant plants under LD. The cell sizes were marked with red lines.
(B) and (C) Cell length and cell number in 7-d-old Col-0 and mlk1 mlk2 double mutants under LD. Means ± sd obtained from over 40 independent plants.
(D) Hypocotyl lengths of Col-0, mlk1 mlk2 double mutants, and rga-28 grown on increasing concentrations of GA3 (0, 0.5, 1.0, 2.0, and 5.0 µM) under LD. Means ± sd obtained from over 40 independent plants.
(E) Relative responses of hypocotyl lengths in Col-0, mlk1 mlk2 double mutants, and rga-28 to GA3 treatment under LD. The response of Col-0 to GA3 is considered 100%, and the response of all mutants to GA3 is shown relative to Col-0. Means ± se; n = 3, where n is the number of independent experiments.
(F) Hypocotyl lengths of Col-0, mlk1 mlk2 double mutants, and rga-28 grown on increasing concentrations of PAC (0, 0.01, 0.02, 0.05, 0.1, 0.2, and 0.5 µM) under LD. Means ± sd obtained from over 40 independent plants.
(G) Relative responses of hypocotyl lengths in Col-0, mlk1 mlk2, and rga-28 to PAC treatment under LD. The response of Col-0 to PAC is considered 100%, and the response of all mutants to PAC is shown relative to Col-0. Means ± se; n = 3, where n is the number of independent experiments.
MLK1 and MLK2 Interact with RGA
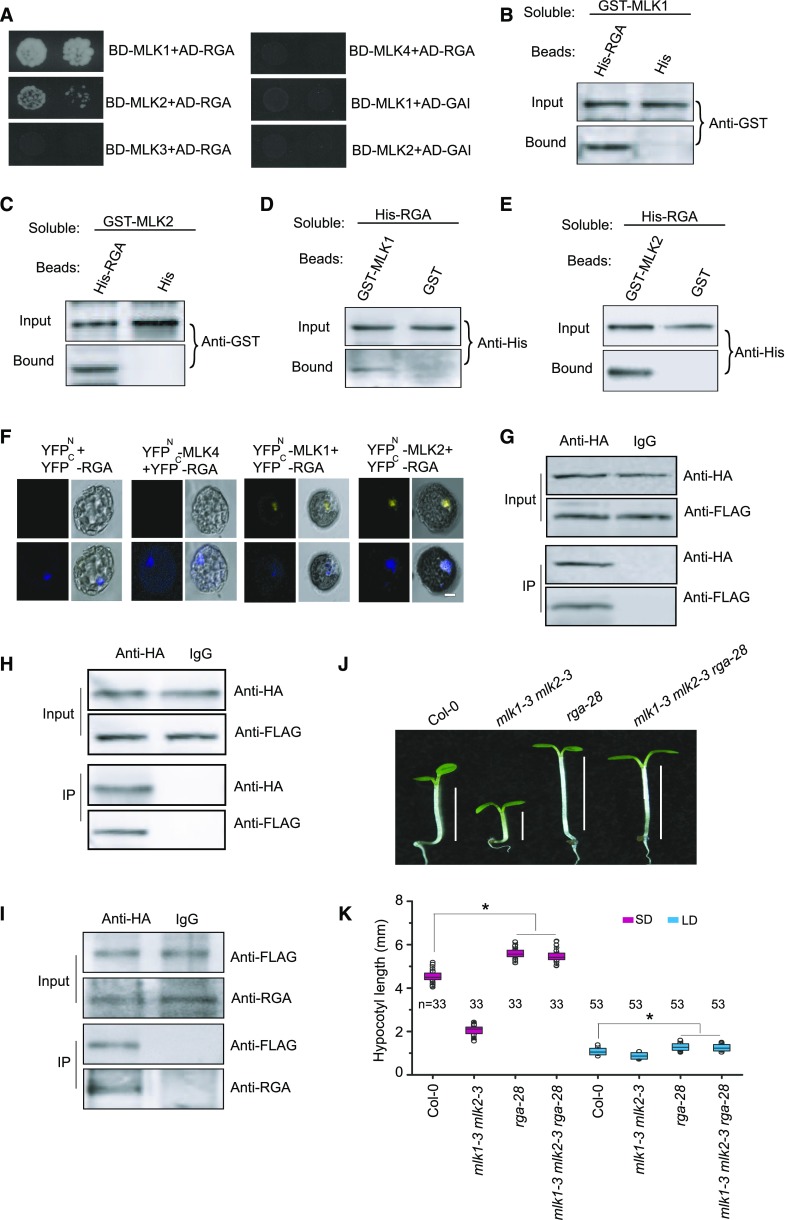
To elucidate the GA signaling network involving MLK1 and MLK2, we screened for proteins that interact with MLK1 and MLK2. We performed yeast two-hybrid analysis with MLK1 and MLK2 fused with the DNA binding domain to identify proteins involved in GA signaling, which were fused to the activation domain. RGA, but not GAI, RGL1, RGL2, or RGL3, interacted with MLK1 and MLK2. Neither MLK3 nor MLK4 interacted with RGA (Figure 3A). The interaction between MLK1/2 with RGA was confirmed by a protein pull-down assay. Proteins containing MLK1 and MLK2 fused with GST bound to beads containing a His-tag fused to RGA, but not to His-tagged beads alone (Figures 3B and 3C). In a complementary experiment, beads attached to GST-MLK1 and GST-MLK2, but not the GST control, bound to soluble His-tagged RGA (Figures 3D and 3E).
Figure 3.
MLK1 and MLK2 Interact with RGA.
(A) Yeast two-hybrid analysis revealing an interaction between MLK1/2 and RGA. The growth of two concentrations (2 × 10−2 and 2 × 10−3) of yeast cultured on synthetic defined medium lacking Trp, Leu, His, and adenine is shown.
(B) and (C) Pull-down assays with MLK1/2 and RGA. Beads containing His-tag (His) or His-fused RGA were assayed for their ability to bind soluble GST-fused MLK1/2. The input or bound protein was detected using an anti-GST antibody.
(D) and (E) Reciprocal pull-down assays with MLK1/2 and RGA. Beads containing GST tag or GST-fused MLK1/2 were assessed for their ability to bind soluble His-fused RGA and detected with an anti-His antibody.
(F) BiFC with MLK1/2 and RGA. MLK1/2 fused to the N terminus of YFP or MLK4 fused to the N terminus of YFP or the N terminus of YFP alone were tested for their ability to bind to the C terminus of YFP fused to RGA. Yellow fluorescence and a bright-field image were recorded and the resulting images were merged. Twenty-five cells were examined for each transformation. Bar = 10 µm.
(G) and (H) Co-IP of MLK1/2 and RGA. FLAG-MLK1/2 and HA-RGA were cotransformed into Arabidopsis protoplasts, immunoprecipitated using an anti-HA antibody, and detected with anti-FLAG and anti-HA antibodies. The cells were harvested at 2 h after lights-on zeitgeber time (ZT2).
(I) Co-IP of MLK2 and RGA in complemented plants. The cell extracts from 10-d-old seedlings were immunoprecipitated using an anti-FLAG antibody and detected with anti-FLAG and anti-RGA antibodies. The seedlings were harvested at 2 h after lights-on zeitgeber time (ZT2).
(J) Representative images of Col-0, rga-28, mlk1 mlk2, and mlk1 mlk2 rga28 under SD conditions. The hypocotyl lengths are indicated with lines.
(K) Hypocotyl lengths of Col-0, rga-28, mlk1 mlk2, and mlk1 mlk2 rga28 plants under SD and LD. Means ± sd obtained from independent plants. Asterisks indicate significant difference using Student’s t test (P < 0.05).
This pull-down interaction was confirmed by bimolecular fluorescence complementation (BiFC). We observed functional YFP in the nucleus after coexpression of MLK1/2-YFPN (fused with the N-terminal half of yellow fluorescent protein) and RGA-YFPC (fused with the C-terminal half of YFP) in Arabidopsis protoplasts, but not in the controls, providing further evidence that MLK1/2 bind directly to RGA (Figure 3F). The MLK1/2-RGA interaction was further validated by coimmunoprecipitation (co-IP) assays. FLAG-MLK1 or FLAG-MLK2 was cotransformed with HA-RGA into Arabidopsis protoplasts, followed by immunoprecipitation using anti-HA antibody. MLK1/2, but not the control, bound to RGA (Figures 3G and 3H).
To investigate whether MLK2 could bind RGA in cells, we generated a construct containing the MLK2 native promoter driving MLK2 tagged with FLAG (ProMLK2:FLAG-MLK2) and transformed this construct into the mlk1-3 mlk2-3 double mutant. ProMLK2:FLAG-MLK2 rescued the short-hypocotyl phenotype of mlk1-3 mlk2-3, indicating the fusion protein retains function (Supplemental Figure 2). Cell extracts from 10-d-old seedlings were immunoprecipitated using anti-FLAG antibody and then detected with anti-RGA antibody. MLK2, but not control, bound to RGA (Figure 3I). These results suggested that MLK1 and MLK2 interact with RGA in vitro and in vivo.
To uncover the genetic relationship between MLK1, MLK2, and RGA, we generated the mlk1-3 mlk2-3 rga-28 triple mutant by crossing rga-28 with mlk1-3 mlk2-3. The hypocotyl length in the mlk1-3 mlk2-3 rga-28 triple mutant was similar to that of rga-28 (Figures 3J and 3K), suggesting that MLK1 and MLK2 act in the same pathway as RGA.
MLK1 and MLK2 Interact with CCA1
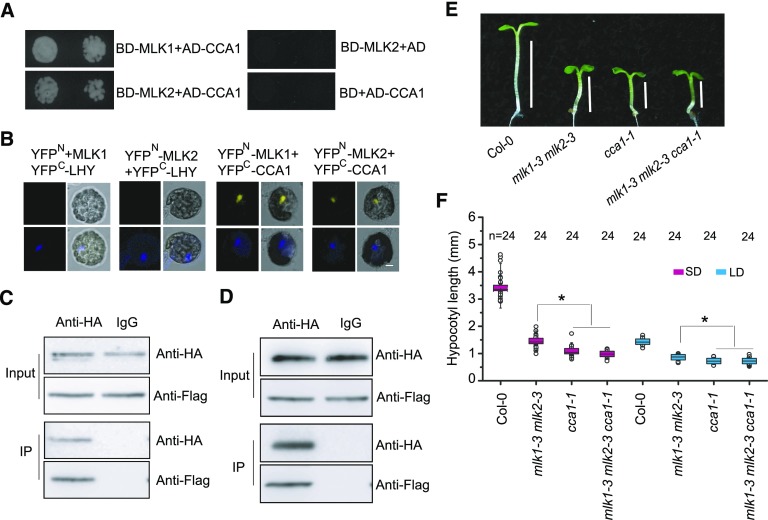
Our recent study showed that CCA1 interacts with MLK4 to regulate flowering time (Su et al., 2017); we therefore examined the interaction between MLK1/2 and CCA1 via a yeast two-hybrid assay. We tested the interaction between MLK1 or MLK2 fused with the binding domain and activation domain-tagged CCA1, finding that both MLK1 and MLK2 interacted with CCA1 (Figure 4A). These yeast two-hybrid interactions were validated by BiFC. Functional YFP was detected in the nucleus of cells cotransformed with MLK1/2-YFPN and CCA1-YFPC, but not in the control cotransformed with MLK1/2-YFPN and LHY-YFPC (Figure 4B). These interactions were further confirmed by co-IP. We observed binding between MLK1/2 and CCA1 in Arabidopsis protoplasts cotransformed with FLAG-MLK1/2 and HA-CCA1, followed by immunoprecipitation with anti-HA antibody (Figures 4C and 4D). These results indicate that MLK1/2 interact with CCA1 in vitro and in vivo.
Figure 4.
MLK1 and MLK2 Interact with CCA1.
(A) Yeast two-hybrid analysis revealing an interaction between MLK1/2 and CCA1. The growth of two concentrations (2 × 10−2 and 2 × 10−3) of yeast cultured on synthetic defined medium lacking Trp, Leu, His, and adenine is shown.
(B) MLK1/2 fused to the N terminus of YFP (YFPN) or the N terminus of YFP alone were tested for their ability to bind to the C terminus of YFP (YFPC) fused to LHY or the C terminus of YFP fused to CCA1. Yellow fluorescence and a bright-field image were recorded and the resulting images were merged. Twenty-five cells were examined for each transformation. Bar = 10 µm.
(C) and (D) Co-IP between MLK1/2 and CCA1. FLAG-MLK1/2 and HA-CCA1 were cotransformed into Arabidopsis protoplasts, immunoprecipitated using an anti-HA antibody, and detected with anti-Flag and anti-HA antibodies. The cells were harvested at 2 h after lights-on zeitgeber time (ZT2).
(E) Representative image of Col-0, cca1-1, mlk1-3 mlk2-3, and cca1-1 mlk1-3 mlk2-3 under SD. Lines indicate hypocotyl lengths.
(F) The hypocotyl lengths of Col-0, cca1, mlk1 mlk2, and cca1 mlk1 mlk2 under SD and LD. Means ± sd obtained from independent plants. Asterisks indicate significant difference using Student’s t test (P < 0.05).
The possibility of a genetic relationship between CCA1 and MLK1/2 was examined by introducing cca1-1 into the mlk1-3 mlk2-3 double mutant background. The hypocotyl length of the cca1 mlk1 mlk2 triple mutant was similar to that of cca1, suggesting that MLK1/2 acts in the same pathway as CCA1 in hypocotyl elongation (Figures 4E and 4F).
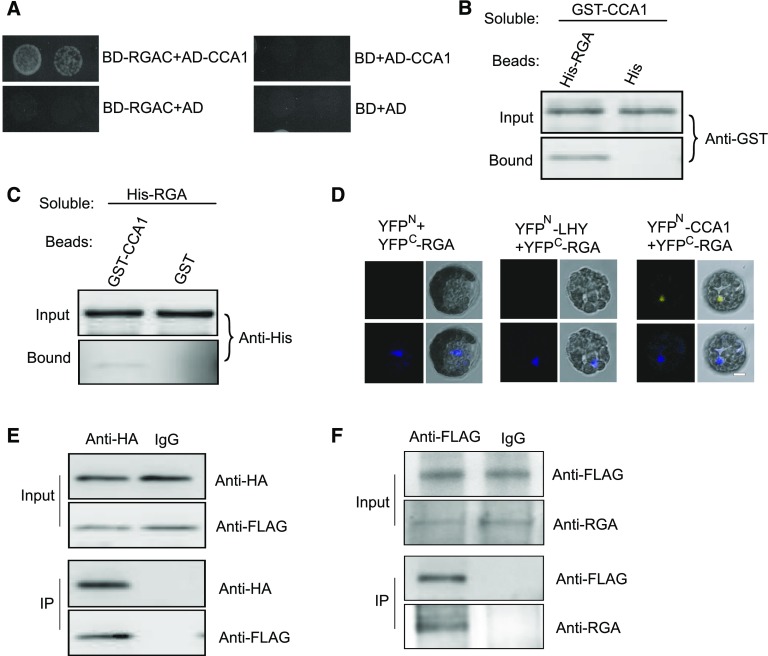
RGA Interacts with CCA1
Given that MLK1/2 physically interact with RGA and CCA1, we investigated the possibility of interaction between RGA and CCA1. Because the N terminus of RGA has autoactivation activity (de Lucas et al., 2008), we used the C terminus of RGA (amino acids 120–587) fused to binding domain to examine this interaction. The C terminus of RGA, but not the control, interacted with CCA1 fused with the activation domain (Figure 5A). This yeast two-hybrid interaction was confirmed by a pull-down assay: Beads containing His fused with full-length RGA, but not the His control, bound to soluble GST-tagged CCA1 (Figure 5B). In a complementary experiment, beads attached to GST-tagged CCA1, but not the GST control, bound to His-tagged RGA (Figure 5C).
Figure 5.
RGA Interacts with CCA1.
(A) Yeast two-hybrid analysis revealing an interaction between the C terminus of RGA and CCA1. The growth of two concentrations (2 × 10−2 and 2 × 10−3) of yeast cultured on synthetic defined medium lacking Trp, Leu, His, and adenine is shown.
(B) Beads containing His tag (His) or His-fused full-length RGA were assayed for their ability to bind soluble GST-fused CCA1. The input and bound proteins were detected with an anti-GST antibody.
(C) Beads containing GST tag or GST-fused CCA1 were assessed for their ability to bind soluble His-fused RGA and detected with an anti-His antibody.
(D) CCA1 fused to the N terminus of YFP (YFPN) or LHY fused to the N terminus of YFP or the N terminus of YFP alone were tested for their ability to bind to the C terminus of YFP (YFPC) fused to full-length RGA. Yellow fluorescence and a bright field image were recorded and the resulting images were merged. Twenty-five cells were examined for each transformation. Bar = 10 µm.
(E) Co-IP between CCA1 and full-length RGA. FLAG-CCA1 and HA-RGA were cotransformed into Arabidopsis protoplasts, immunoprecipitated using an anti-HA antibody, and detected with anti-Flag and anti-HA antibodies. The cells were harvested at 2 h after lights-on zeitgeber time (ZT2).
(F) Co-IP of CCA1 and RGA in complemented plants. The cell extracts from 10-d-old seedlings were immunoprecipitated using an anti-FLAG antibody and detected with anti-FLAG and anti-RGA antibodies. The seedlings were harvested at 2 h after lights-on zeitgeber time (ZT2).
This interaction was further validated by BiFC and co-IP assays. Functional YFP was observed in the nucleus in cells cotransformed with CCA1-YFPN and full-length RGA-YFPC (Figure 5D). Immunoprecipitation using HA antibody revealed that CCA1 bound to RGA in Arabidopsis protoplasts cotransformed with HA-CCA1 and FLAG-RGA (Figure 5E).
We isolated two additional cca1 mutants, cca1-21 and cca1-22, and showed that these mutants lack full-length CCA1 transcript, indicating that they are likely null alleles (Su et al., 2017). The short-hypocotyl phenotype of cca1-22 was rescued by transformation with a construct in which the native CCA1 promoter drives CCA1 tagged with FLAG (ProCCA1:FLAG-CCA1) (Su et al., 2017). We then investigated the interaction between CCA1 and RGA in complemented plants. Cell extracts from 10-d-old seedlings were immunoprecipitated using anti-FLAG antibody and then detected with anti-RGA antibody. CCA1, but not the IgG control, bound to RGA (Figure 5F). These results indicate that RGA interacts with CCA1 in vitro and in vivo.
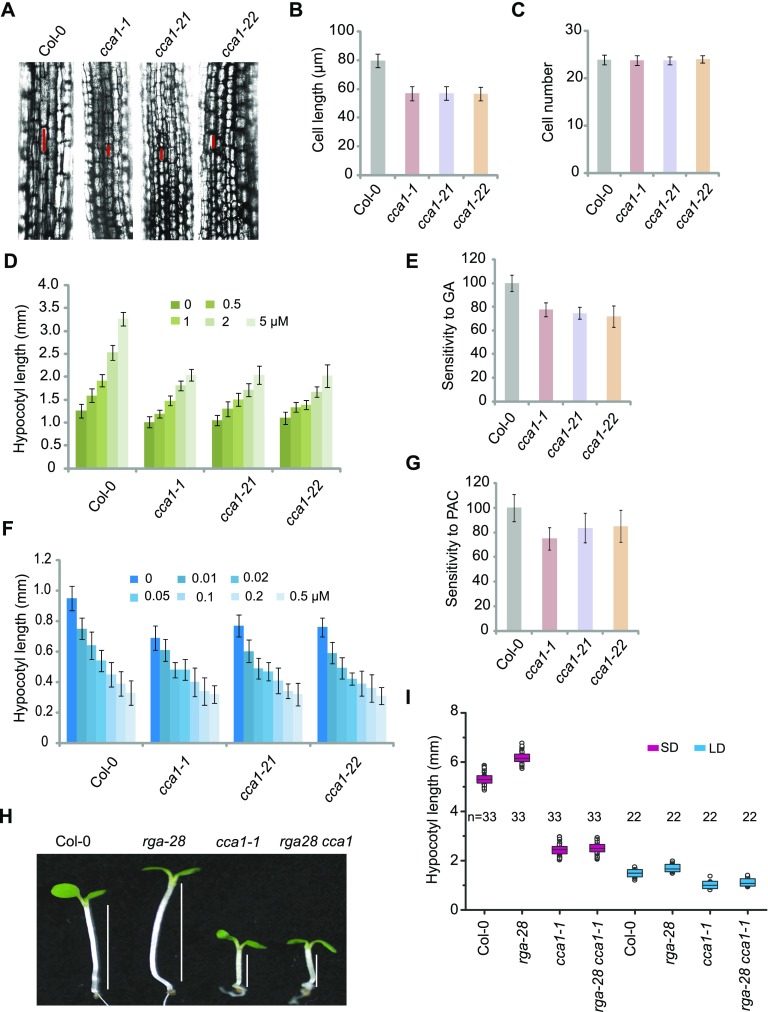
The cca1 Mutants Are Hyposensitive to GA
Examination of hypocotyl lengths, cell lengths, and cell numbers in wild-type and cca1 plants showed that cca1 mutants had shorter hypocotyls than the wild type (Figure 6A). These short hypocotyls were caused by reduced cell length, not by reduced cell number (Figures 6B and 6C).
Figure 6.
Mutations in CCA1 Result in Hyposensitivity to GAs.
(A) Cell lengths in hypocotyls of 1-week-old Col-0 and cca1 plants under LD. The cell sizes are marked with red lines.
(B) and (C) Cell length and cell number in 1-week-old Col-0 and cca1 plants under LD. Means ± sd obtained from over 40 independent plants. Values are the means of three independent experiments; error bars, sd between experiments.
(D) Hypocotyl lengths of Col-0 and cca1 plants grown on increasing concentrations of GA3 (0, 0.5, 1.0, 2.0, and 5.0 µM) under LD. Means ± sd obtained from over 40 independent plants.
(E) Relative responses of hypocotyl lengths in Col-0 and cca1 to GA3 treatment. Means ± se, n = 3, where n is the number of independent experiments.
(F) Hypocotyl lengths of seedlings grown on increasing concentrations of PAC (0, 0.01, 0.02, 0.05, 0.1, 0.2, and 0.5 µM) under LD. Means ± sd obtained from over 40 independent plants.
(G) Relative responses of hypocotyl lengths in Col-0 and cca1 to PAC treatment. Means ± sd obtained from over 40 independent plants.
(H) Representative images of Col-0, rga-28, cca1-1, and rga-28 cca1-1 plants under SD. Lines indicate hypocotyl lengths.
(I) Hypocotyl lengths of Col-0, rga-28, cca1-1, and rga-28 cca1-1 plants under SD and LD. Means ± sd obtained from independent plants. Asterisks indicate significant difference using Student’s t test (P < 0.05).
We then investigated the role of CCA1 in the GA pathway. In the presence of exogenous GA, the rate of elongation and final hypocotyl length increased in wild-type and cca1 plants. Compared with the wild type, the cca1 mutants exhibited a reduced GA response (Figures 6D and 6E). We further investigated the role of GAs in regulating hypocotyl elongation using PAC treatment. The cca1 mutants were less sensitive to PAC than the wild type (Figures 6F and 6G). These results suggest that CCA1 is required for hypocotyl elongation promoted by GAs.
To elucidate the relationship between CCA1 and RGA, we generated the cca1-1 rga-28 double mutant. The hypocotyl length of the double mutant was similar to that of cca1 (Figures 6H and 6I), indicating that CCA1 is epistatic to RGA.
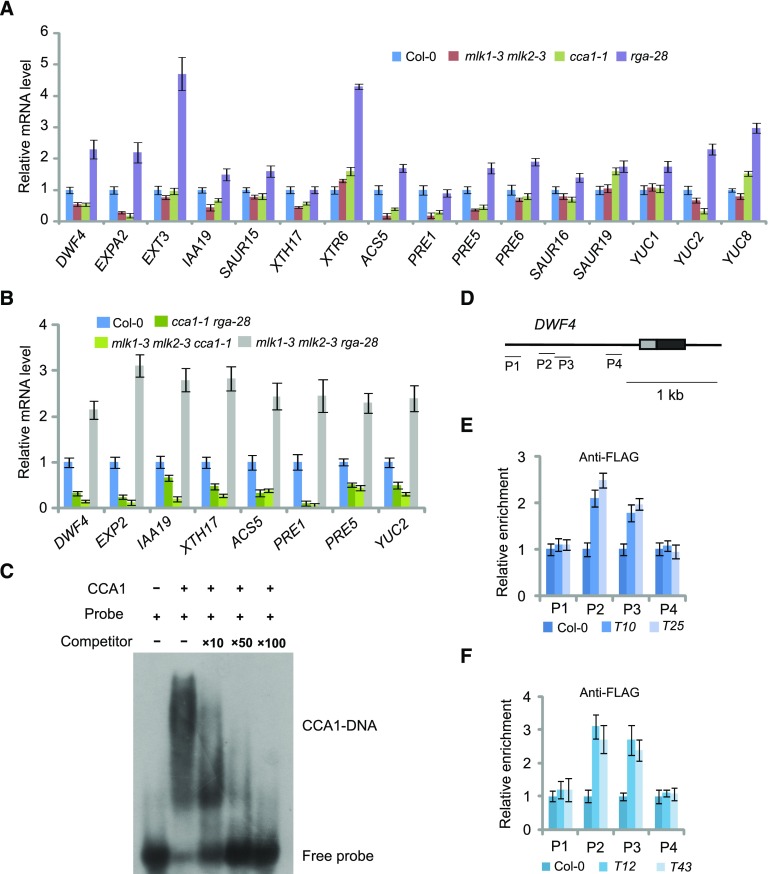
CCA1 and MLK1/2 Directly Target DWF4
Given that MLK1/2 physically interact with RGA and CCA1, we investigated whether any genes involved in cell expansion are coregulated by MLK1/2, CCA1, and RGA. The transcript levels of cell elongation-related genes, including DWF4, EXPANSIN A2 (EXPA2), EXTENSIN3, INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE7 (XTH17), XTR6, PACLOBUTRAZOL RESISTANCE1 (PRE1), PRE5, PRE6, SAUR-LIKE AUXIN-RESPONSIVE PROTEIN FAMILY (SAUR15), SAUR16, SAUR19, ACC SYNTHASE5 (ACS5), YUCCA1 (YUC1), YUC2, and YUC8, were measured by quantitative RT-PCR using tissue from wild type, cca1-1, rga-28, and mlk1-3 mlk2-3 seedlings. The transcript levels of DWF4, EXPA2, IAA19, XTH17, ACS5, PRE1, PRE5, and YUC2 were reduced in cca1-1 and mlk1 mlk2 plants, but increased in rga-28 (Figure 7A), indicating that these genes are activated by CCA1 and MLK1/2, but suppressed by RGA. These gene expression patterns are consistent with the hypocotyl measurements described above, suggesting that CCA1, MLK1/2, and RGA coordinately modulate hypocotyl elongation. To confirm these results, we measured the transcript levels of DWF4, EXPA2, IAA19, XTH17, ACS5, PRE1, PRE5, and YUC2 in cca1-1 rga-28, mlk1-3 mlk2-3 rga-28, and mlk1-3 mlk2-3 cca-1 mutants. The transcript levels of these genes increased in mlk1-3 mlk2-3 rga-28 and decreased in cca1-1 rga-28 and mlk1-3 mlk2-3 cca-1 mutants (Figure 7B).
Figure 7.
CCA1 Binds the Promoters of DWF4.
(A) Relative transcript levels of genes related to cell expansion were measured in Col-0, cca1-1, rga-28, and mlk1-3 mlk2-3 mutants. Experiments were repeated at least three times, and each experiment shown indicates the mean ± se, n = 3 replicates. The leaves was harvested at 2 h after lights-on zeitgeber time (ZT2) and used for RT-PCR.
(B) Relative transcript levels of genes related to cell expansion were measured in Col-0, cca1-1rga-28, mlk1-3 mlk2-3 cca1-1, and mlk1-3 mlk2-3 rga-28 plants. Experiments were repeated at least three times, and each experiment shown indicates the mean ± se, n = 3 replicates. The leaves were harvested at 2 h after lights-on zeitgeber time (ZT2) and used for RT-PCR.
(C) Gel shift assay with CCA1 and fragments of the DWF4 promoter region, respectively. The binding ability of CCA1 to fragments of the DWF4 promoter labeled with 32P was assessed, and this binding specificity was tested by adding unlabeled competitor probe.
(D) Diagram of the DWF4 promoter including untranslated regions (gray boxes), exons (black boxes), and introns (lines). The tested regions are marked with lines. The CCA1 binding sites are located between region P2 and P3.
(E) and (F) The relative enrichment of CCA1(E) and MLK2 (F) at the DWF4 promoter was assessed with ChIP-PCR. The cca1 complemented with ProCCA1:FLAG-CCA1 were indicated as T10 and T25, and mlk2 complementary plants (T12 and T43) are described in Supplemental Figure 2. Experiments were repeated at least three times, and each experiment shown indicates the mean ± se, n = 3 replicates. The leaf was cross-linked at 2 h after lights-on zeitgeber time (ZT2).
Next, we examined the promoter sequences of these genes and asked whether CCA1 targets these genes directly. Sequence analysis identified the conserved CCA1 binding motif, AAATATCT (Nagel et al., 2015), in the promoter of DWF4 (Supplemental Figure 3). We therefore examined the direct binding of CCA1 to fragments of the DWF4 promoter by electrophoretic mobility shift assay (EMSA). The EMSA showed retarded bands in the presence of CCA1, but not when the reaction included specific competitor probes (Figure 7C).
To test whether CCA1 bound the promoter of DWF4 in vivo, we examined CCA1 occupancy using two cca1 mutants harboring the ProCCA1:FLAG-CCA1 transgene (T10 and T25), which rescued the short hypocotyls of cca1 (Su et al., 2017). The profiles of CCA1 were measured by chromatin immunoprecipitation (ChIP) with a FLAG-specific antibody, followed by quantitative PCR analysis of the amount of DNA enrichment. Strong enrichment of CCA1 was observed in regions 2 and 3 of DWF4 in two complemented plants (Figures 7D and 7E). These results indicated that CCA1 binds the promoter of DWF4 in vitro and in vivo.
Given that CCA1 interacts with MLK1/2 and binds to the DWF4 promoter to directly regulate DWF4, it is possible that MLK1/2 might also regulate DWF4 directly. We investigated MLK2 enrichment in the DWF4 promoter region using complemented plants harboring ProMLK2:FLAG-MLK2 (Supplemental Figure 2). The profiles of MLK2 were measured using ChIP-PCR with a FLAG-specific antibody. This showed that MLK2 was enriched at the promoter of DWF4 (Figures 7D and 7F), indicating that MLK2 directly regulates DWF4.
The transcripts of MLK1/2 were then investigated. The RT-PCR results revealed that MLK1/2 were repressed at ZT4 and induced at ZT16 (Supplemental Figures 4A and 4B). Since CCA1 is unstable and CCA1 directly targets DWF4, we investigated the transcripts of DWF4 and found that DWF4 was slightly activated at ZT16 (Supplemental Figure 4C).
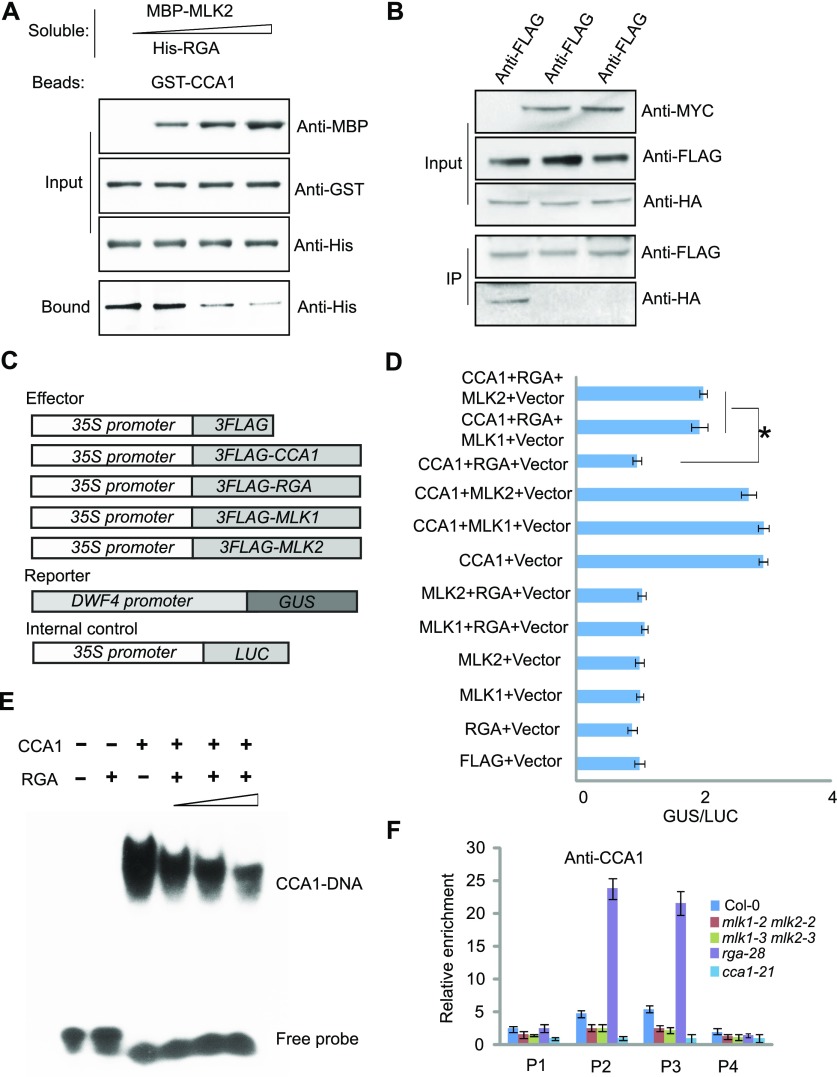
MLK1/2 Antagonize the Function of RGA to Interact with CCA1
Since MLK1/2 are homologs of MUT9p, we assessed the phosphorylation activity of MLK1/2 with RGA and CCA1 as substrates. MLK1/2 phosphorylated H3, but not RGA, or CCA1 (Supplemental Figure 5), suggesting that MLK1/2 might not function as a kinase for RGA and CCA1, and not be involved in posttranslational regulation of RGA and CCA1. We then investigated whether any phosphorylation of H3 at threonine 3 (H3T3ph) was reduced in mlk1 mlk2 double mutants with ChIP-PCR because mutations in MLK1 and MLK2 result in a reduction of phosphorylation of H3 at threonine 3 (Wang et al., 2015). The enrichment of H3T3ph at DWF4 was not notable between the wild type and mlk1 mlk2 mutants, suggesting that MLK1/2 regulate DWF4 independently of H3T3 phosphorylation (Supplemental Figure 6).
As observed above, MLK1/2 physically interact with RGA and CCA1, and the function of MLK1/2 in hypocotyl elongation via GA signal transduction is opposite to that of RGA. Therefore, we investigated whether MLK1/2 and RGA antagonistically interact with CCA1. We investigated the antagonistic functions of MLK1/2 and RGA versus CCA1 via competitive pull-down assays. The interaction between RGA and CCA1 decreased with increasing MLK2 level, indicating that MLK2 and RGA antagonistically interact with CCA1 (Figure 8A). To confirm these results, we cotransformed Arabidopsis protoplasts with HA-RGA and FLAG-CCA1 with or without MYC-MLK1/2, followed by co-IP with anti-FLAG and detection with anti-HA. RGA was enriched in the absence of MLK1/2, but lower levels were detected in the presence of MLK1/2. We therefore conclude that MLK1/2 and RGA antagonistically interact with CCA1 (Figure 8B).
Figure 8.
MLK2 Antagonizes the Function of RGA to Bind CCA1 in Vitro and in Vivo.
(A) Competitive pull-down of CCA1 with RGA and MLK2. Beads containing GST-fused CCA1 were assayed for their ability to bind a soluble His-fused RGA with increased dosage of MBP-MLK2.
(B) Co-IP between RGA and CCA1 with or without MLK1/2. FLAG-CCA1 and HA-RGA were cotransformed with or without MYC-MLK1/2 into Arabidopsis protoplasts, immunoprecipitated using an anti-FLAG antibody, and then detected with anti-Flag and anti-HA. The cells were harvested at 2 h after lights-on zeitgeber time (ZT2).
(C) The vectors used in the GUS activity assay.
(D) The GUS activity from the ProDWF4:GUS reporter construct for cells transformed with CCA1, RGA, and MLK1/2. The x axis shows the relative GUS activity compared with the internal luciferase control (35S:LUC). Asterisks indicate P < 0.05 by t test.
(E) Gel shift assay with CCA1 and fragments of the DWF4 promoter region with increasing dosage of RGA. The binding ability of CCA1 to fragments of the DWF4 promoter labeled with 32P was assessed with increasing amounts of RGA (1, 4, and 10 µg).
(F) The relative enrichment of CCA1 at the DWF4 promoter was assessed using ChIP-PCR in rga28 and mlk1 mlk2 mutants. Experiments were repeated at least three times, and each experiment shown indicates the mean ± se, n = 3 replicates. The leaves were cross-linked at 2 h after lights-on zeitgeber time (ZT2).
We next examined whether this antagonistic interaction occurs at the promoter of DWF4, which is a target of CCA1 and MLK2. ProDWF4:GUS was transformed into protoplasts with HA-CCA1 or vector alone. We detected high levels of GUS activity in cells cotransformed with CCA1 compared with vector alone, suggesting that CCA1 promotes the transcription of DWF4. Then, HA-RGA and HA-MLK1/2 were sequentially added in the GUS reporter system. GUS activity was highly induced in the cells cotransformed with CCA1 and MLK1/2 but was suppressed in the cells cotransformed with CCA1 and RGA. The repression of GUS activity by RGA was reversed in the presence of CCA1, RGA, and MLK1/2 (Figures 8C and 8D), suggesting that MLK1/2 antagonize RGA to modulate the activity of CCA1. These results are consistent with the hypocotyl lengths of cca1, rga, and mlk1 mlk2 seedlings and the expression level of DWF4 in these different backgrounds. We then used EMSA to investigate whether RGA can inactivate CCA1 by blocking CCA1 binding to DWF4. The EMSA showed retarded bands in the presence of CCA1, but these bands decreased in intensity with increasing dosage of RGA (Figure 8E), suggesting that RGA inactivates CCA1 binding affinity.
The antagonistic interaction of MLK1/2 and RGA with CCA1 was further measured with ChIP-PCR. The CCA1 that bound to the DWF4 promoter in rga-28 and mlk1 mlk2 double mutants was immunoprecipitated with CCA1-specific antibody, followed by analysis of the amount of DNA with quantitative PCR. The enrichment of CCA1 at the DWF4 promoter was reduced in mlk1 mlk2 double mutants, but increased in rga-28 (Figure 8F), thereby providing further evidence that CCA1 binding was antagonistically competed by MLK1/2 and RGA.
DISCUSSION
In this study, we demonstrated that MLK1/2, RGA, and CCA1 coordinate plant development, thus providing insight into the mechanism by which the circadian clock protein CCA1 combines with RGA and MLK1/2 to regulate hypocotyl length. The Arabidopsis genome contains four genes encoding MUT9p-like proteins: MLK1, MLK2, MLK3, and MLK4 (Wang et al., 2015; Huang et al., 2016). We found that MLK1 and MLK2, but not MLK3 or MLK4, interact with RGA, suggesting that MUT9p evolved into MLK1 and MLK2, which have divergent functions in Arabidopsis. These results were also supported by the phenotypes of the mlk1 mlk2 double mutants. Loss of MLK1 and MLK2 function led to hyposensitivity to GA, resulting in short hypocotyls due to reduced cell length. The function of MLK1/2 in the GA pathway was further supported by the finding that the mlk1-3 mlk2-3 rga-28 triple mutant displayed long hypocotyls, like rga-28. MLK1/2 and CCA1 promote hypocotyl elongation, but RGA represses this process, suggesting that MLK1/2 might prevent RGA from binding to specific genes. These results were supported by the finding that genes involved in cell elongation are mostly regulated by MLK1/2 and RGA in an opposite manner. MLK1/2 interact with RGA and antagonistically bind to CCA1 in vitro and in vivo. CCA1 increased the activity of a GUS reporter under the control of the DWF4 promoter, but RGA suppressed GUS expression. The decrease in GUS activity by RGA was reversed by MLK1/2, suggesting that MLK1/2 antagonize RGA to modulate the activation of DWF4 expression by CCA1. The deposition of CCA1 was enriched in rga-28, but reduced in mlk1 mlk2 double mutants, further providing evidence that MLK1/2 antagonize RGA in vivo.
MLK1/2 were first identified as kinases of histone H3 at threonine 3 and are involved in the osmotic stress response (Wang et al., 2015), but in our study, MLK1/2 were not observed to affect the phosphorylation of histone H3 at threonine 3 at DWF4. MLK1/2 physically interact with RGA, but not the other DELLA proteins, suggesting that MLK1/2 might be involved specifically in the regulation of hypocotyl elongation by GA. Both MLK1/2 and rice EL1 encode casein kinase I, and these proteins show high similarity to yeast YCK2 and CKI (Dai and Xue, 2010; Wang et al., 2015; Huang et al., 2016). EL1 was observed to phosphorylate the DELLA protein SLR1. Loss of EL1 function results in early flowering, and overexpression of EL1 leads to a dwarf phenotype via increased SLR1 accumulation (Dai and Xue, 2010). By contrast, loss of MLK1 and MLK2 function results in late flowering and short hypocotyl length, suggesting that Arabidopsis MLK1/2 might have evolved divergent functions from rice EL1. MLK1/2 is a component of the evening complex of the circadian clock and phytochrome B (Huang et al., 2016). Phytochrome B not only mediates the red light-induced inhibition of stem growth, but also regulates seedling responsiveness to GAs (Reed et al., 1996; Olszewski et al., 2002), suggesting that MLK1/2 might be involved in multiple signaling pathways in the developmental process.
DELLAs are important integrators of signals from other phytohormones and environmental factors. DELLAs were observed to bind and inactivate transcription factors (de Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010; Bai et al., 2012), and the stability of DELLAs was modulated by translational modifications, suggesting that DELLAs are vital factors that are subject to complex regulation in plant development and defense. Genes that function in plant growth and cell expansion are coregulated by DELLAs and circadian clock-controlled processes (Arana et al., 2011), suggesting that DELLAs and the circadian clock are involved in the same processes during plant growth. Circadian clock gates GA signaling through transcriptional regulation of the GA receptors, resulting in higher stability of DELLA proteins during daytime and higher GA sensitivity at night, and oscillation of GA signaling appears to be particularly critical for rhythmic growth (Arana et al., 2011), suggesting that circadian clock and GA signaling crosstalk in multilevel. Our study showed that RGA physically interacts with CCA1 and inactivates the binding of CCA1 to DWF4, suggesting that this DELLA protein directly integrates the circadian clock and GA pathways to regulate plant development.
Loss of function of CCA1 results in short hypocotyls and overexpression of CCA1 results in long hypocotyls (Wang and Tobin, 1998; Niwa et al., 2007). CCA1 controls the photoperiodic response of hypocotyl elongation by modulating the rhythmic expression of PIF4 and PIF5 (Niwa et al., 2009). PIF4 and PIF5 interact with DELLAs to coordinate light and GA signaling during cell elongation (de Lucas et al., 2008; Feng et al., 2008), suggesting that CCA1 and DELLAs regulate hypocotyl elongation via multilevel interactions among the circadian clock, light, and GAs. Mutations in CCA1 resulted in short hypocotyls, and this short-hypocotyl phenotype is not reversed by exogenous GA application. In addition, cca1-1 rga-28 double mutants have short hypocotyls, like cca1-1 mutants, suggesting that CCA1 promotes hypocotyl elongation via the GA pathway.
The cca1 mlk1 mlk2 triple mutant displays the short hypocotyl phenotype, which is similar to cca1, suggesting that MLK1/2 act in the same pathway as CCA1. Although our statistical analysis showed that the difference in hypocotyl length between mlk1 mlk2 and cca1 mlk1 mlk2 was significant, the difference was reduced under LD. Light inhibits elongation of mlk1 mlk2 and cca1 mlk1 mlk2 hypocotyls, which in turn minimizes the difference of between cca1mlk1 mlk2 and mlk1 mlk2. Collectively, our study demonstrates that GAs and the circadian clock regulate plant development via a process modulated by MLK1/2.
METHODS
Plant Materials
Arabidopsis thaliana ecotype Col-0 plants were grown at 22°C under a LD photoperiod (16-h-light/8-h-dark cycle) and light intensity of 160 µmol m−2 s−1 (T50 fluorescent lamp; Philips) or a SD photoperiod (8-h-light/16-h-dark cycle). The mutant strains from the SALK collection were as follows: mlk1-2, Salkseq_132455; mlk1-3, SALK_039903; mlk2-2, SALK_149222; mlk2-3, SALK_064333; cca1-21, SALKseq123282; cca1-22, SALKseq120169; and rga-28, SALK_089146. Seeds from the cca1-1 mutant, which had been backcrossed to Col-0 for six generations, were a gift from Hongtao Liu of the Institute of Physiology and Ecology, SIBS, CAS.
Plasmid Constructs
The plasmids were constructed using the DNA primers and protocols described in Supplemental File 1. All cloned DNAs were confirmed by DNA sequencing.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed according to the manufacturer’s protocol (Clontech user manual 630489). Briefly, the Saccharomyces cerevisiae strain AH109 was transformed with the bait constructs pGBKT-MLK1, pGBKT-MLK2, or pGBKT-RGAC and then transformed with pGADT7-RGA, pGADT7-GAI, pGADT7-RGL1, or pGADT7-CCA1. Vectors lacking coding region insertions were used as negative controls. The yeast was scored for protein interaction based on their ability to grow on synthetic defined medium lacking Trp, Leu, His, and adenine. The primers used to generate the constructs are shown in Supplemental File 1.
Hypocotyl Length Analyses
Seeds were surface sterilized and grown on vertical plates with 0.5× Murashige and Skoog medium at 22°C under LD, SD, or darkness. For hypocotyl length, 7-d-old seedlings were measured using microscope systems (Leica; M165C). Mean values were used to calculate the difference between mutant and wild-type seedlings. P values were determined by Student’s t test. The box plot was generated with Origin and the column was generated with Excel.
For GAs and PAC treatment, the seeds were grown on 0.5× Murashige and Skoog medium containing different concentration of GA3 (Sigma-Aldrich) or PAC (Sigma-Aldrich) at 22°C for 7 d. The hypocotyl lengths were measured and mean values were used to calculate the difference between mutant and wild-type seedlings and the relative differences between mutant and wild-type seedlings (wild-type values set to unity).
Transient Expression in Arabidopsis Protoplasts and BiFC
For BiFC, MLK1, MLK2, CCA1, and RGA were cloned into pUC-SPYCE (amino acids 156–239) or pUC-SPYNE (amino acids 1–155) (Walter et al., 2004). Arabidopsis mesophyll protoplasts were isolated and transformed as described previously (Yoo et al., 2007; Lu et al., 2017). Briefly, the leaves of 3-week-old Arabidopsis plants were detached and placed onto double-sided tape, digested with enzymes, and washed with MMG buffer (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7). The protoplasts were cotransformed with the corresponding constructs and examined under a confocal laser scanning microscope (Zeiss LSM710) or immunoprecipitated with specific antibodies.
Protein Pull-Down Assays, Co-IP, and Immunoblot Assays
For the pull-down assays, beads were incubated with 3 µg of fusion protein, washed, and incubated with 3 µg of soluble protein overnight at 4°C. Mock controls included extracts prepared from either the His-Tag or GST vectors. The beads were washed five times with a solution containing 20 mM Tris (pH 7.4), 150 mM NaCl, and 0.05% Tween 20, separated on an SDS-PAGE gel, and analyzed by immunoblotting using an anti-GST antibody (GenScript; A00866-100, lot 13D000626) or an anti-His antibody (Abmart; M30111M, lot 273884).
For the co-IP, MLK1/2, CCA1, and RGA were fused with FLAG, HA, or MYC and cloned into the pUC19 vector. Co-IP was performed as described previously (Zhang et al., 2015; Lu et al., 2017). Briefly, 1 × 106 Arabidopsis protoplasts were lysed with PEN-140 buffer (140 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA, and 0.05% CA-630). The supernatant was precleared with Protein G and precipitated with anti-FLAG (Sigma-Aldrich; H6908, lot SLBQ7119V) or anti-HA (Sigma-Aldrich; H9658, lot 095M4778V) antibodies. The protein complexes were isolated by binding to Protein G beads, followed by five washes with PEN-400 buffer (400 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA, and 0.05% CA-630). The samples were analyzed by immunoblotting using an anti-MYC (Millipore; 05-724, lot 2762289), anti-RGA (Agrisera; AS11 1630, lot 1511), anti-HA, or anti-FLAG antibody.
Phosphorylation Reaction in Vitro
The phosphorylation reaction assay was performed as described (Demidov et al., 2005; Lu et al., 2017). Briefly, 2 µg of protein was incubated in reaction buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 2 mM EDTA, and 50 µM ATP) with 2.5 µCi [γ-32P]ATP at 30°C for 1 h. The reaction products were separated by SDS-PAGE and autoradiographed using x-ray film.
EMSA
The EMSA was performed as described (Su et al., 2017). Briefly, 1 to 2 µg purified protein was mixed with 4 pmol [γ-32P]ATP-labeled probe with or without various amounts of unlabeled probe. After separation in a 4.5% native nondenaturing acrylamide gel, the gel was exposed to x-ray film overnight. The sequence of the probe is shown in Supplemental File 1.
Quantitative RT-PCR
Total RNA was isolated and reverse transcription was performed with oligo(dT) primers (Promega), and the transcript levels of individual genes were measured using gene-specific primers. RT-PCR analysis was performed with a CFX real-time PCR instrument (Bio-Rad) and SYBR Green mixture (Roche). The relative expression level of the genes was quantitated via the 2−ΔΔCT Ct method using UBIQUITIN as the reference housekeeping gene for expression analysis and for relative measurements of input DNA for the chromatin immunoprecipitation assays. Primer information is provided in Supplemental File 1.
ChIP Assay
ChIP was performed as described (Lu et al., 2017). Briefly, 3 g of 10-d-old seedlings was fixed with 1% formaldehyde for 10 min and quenched in 0.125 M glycine. The leaves were ground in a mortar and pestle in buffer I (0.4 M sucrose, 10 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and filtered through Miracloth. After centrifugation, the pellet was extracted with buffer II (0.25 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and then with buffer III (1.7 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail). The nuclei were then lysed in lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and the extract was sonicated to fragment the DNA to a size range of 300 to 500 bp. After centrifugation at 12,000 rpm for 10 min at 4°C, the supernatant was diluted in dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.0, 167 mM NaCl, 0.1 mM PMSF, and protease inhibitor cocktail) then precleared with protein A or protein G magnetic beads. Specific antibodies (Sigma-Aldrich; H6908, lot SLBQ7119V) or anti-HA (Sigma-Aldrich; H9658, lot 095M4778V), anti-CCA1 (Abiocode; R1234-3, lot 14057), or control IgG serum were added to the precleared supernatants for an overnight incubation at 4°C. The antibody-protein complexes were isolated by binding to protein A or protein G beads. The washed beads were heated at 65°C for 8 h with proteinase K to reverse the formaldehyde cross-linking and digest the proteins. The sample was then extracted with phenol/chloroform, and the DNA was precipitated in ethanol and resuspended in water. The purified DNA was analyzed by real-time PCR with gene-specific primers (Supplemental File 1).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MLK1 (At5g18190), MLK2 (At3g03940), MLK3 (At2g25760), and MLK4 (At3g13670).
Supplemental Data
Supplemental Figure 1. The mlk1 mlk2 double mutants exhibit late flowering.
Supplemental Figure 2. Complementation of the mlk1-3 mlk2-3 double mutant with ProMLK2:FLAG-MLK2.
Supplemental Figure 3. The promoter of DWF4 contains CCA1 binding sites.
Supplemental Figure 4. The transcripts of MLK1, MLK2, and DWF4.
Supplemental Figure 5. The phosphorylation activity of MLK1 and MLK2.
Supplemental Figure 6. The phosphorylation of H3 at threonine 3 in the DWF4 locus.
Supplemental File 1. Plasmids and primers.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 31371306, 31571315, and 91435101 to Y.D.) and the Strategic Priority Research Program “Molecular Mechanism of Plant Growth and Development” of CAS (Grant XDPB04). We thank Suiwen Hou from Lanzhou University for kindly providing transgenic rga-28 seeds, Hongtao Liu from the Institute of Physiology and Ecology, SIBS, CAS for kindly providing cca1-1 seeds, and Ertao Wang from the Institute of Physiology and Ecology, SIBS, CAS for kindly providing pUC18-FLAG and pUC18-HA vectors.
AUTHOR CONTRIBUTIONS
H.Z. and Y.D. conceived the study and designed the experiments. H.Z. performed most of the experiments, and all authors took part in interpreting the results and preparing the manuscript. Y.D. wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Achard P., Renou J.-P., Berthomé R., Harberd N.P., Genschik P. (2008a). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660. [DOI] [PubMed] [Google Scholar]
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008b). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Zhang X., Zhu Z., Ji Y., He W., Jiang Z., Li M., Guo H. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana M.V., Marín-de la Rosa N., Maloof J.N., Blázquez M.A., Alabadí D. (2011). Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 9292–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Murase K., Sun T.P., Steber C.M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R., Wu Y., LoCascio J.C., Feldmann K.A. (1998). An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.-Y., Shang J.-X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.-Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano J.A., Jeong B.R., Xu J., Moriyama H., Cerutti H. (2008). The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. USA 105: 6486–6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D.E., Cao D., Luo D., Harberd N.P., Peng J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064. [DOI] [PubMed] [Google Scholar]
- Choe S., Dilkes B.P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D. (1996). Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 10: 1–8. [DOI] [PubMed] [Google Scholar]
- Cowling R.J., Harberd N.P. (1999). Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J. Exp. Bot. 50: 1351–1357. [Google Scholar]
- Dai C., Xue H.W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29: 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.-M., Achard P. (2013). Gibberellin signaling in plants. Development 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- Davière J.-M., de Lucas M., Prat S. (2008). Transcriptional factor interaction: a central step in DELLA function. Curr. Opin. Genet. Dev. 18: 295–303. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.-M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Demidov D., Van Damme D., Geelen D., Blattner F.R., Houben A. (2005). Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Sun T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Fleck B., Xie D., Burton N., Harberd N.P. (2004). The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K., Sasaki A., Itoh H., Ueguchi-Tanaka M., Ashikari M., Kitano H., Matsuoka M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37: 626–634. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Östin A., Sandberg G., Romano C.P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd N.P., Belfield E., Yasumura Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y.C., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- Huang H., Alvarez S., Bindbeutel R., Shen Z., Naldrett M.J., Evans B.S., Briggs S.P., Hicks L.M., Kay S.A., Nusinow D.A. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15: 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Sasaki A., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Hasegawa Y., Minami E., Ashikari M., Matsuoka M. (2005). Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46: 1392–1399. [DOI] [PubMed] [Google Scholar]
- Lee S., Cheng H., King K.E., Wang W., He Y., Hussain A., Lo J., Harberd N.P., Peng J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16: 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Tian Y., Wang S., Su Y., Mao T., Huang T., Xu Z. (2017). Phosphorylation of SPT5 by CDKD; 2 is required for VIP5 recruitment and normal flowering in Arabidopsis thaliana. Plant Cell 29: 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Webb C.J., Knowles S.M., Kim S.H., Wang Z., Tobin E.M. (2012). CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158: 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R.M., Peters J.M., Graff J.M. (2001). The casein kinase I family: roles in morphogenesis. Dev. Biol. 235: 378–387. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.-R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641. [DOI] [PubMed] [Google Scholar]
- Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S.K., Steber C.M. (2016). Gibberellin hormone signal perception: down‐regulating DELLA repressors of plant growth and development. Annu. Plant Rev. 49: 153–188. [Google Scholar]
- Nieto C., López-Salmerón V., Davière J.-M., Prat S. (2015). ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr. Biol. 25: 187–193. [DOI] [PubMed] [Google Scholar]
- Niwa Y., Yamashino T., Mizuno T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854. [DOI] [PubMed] [Google Scholar]
- Niwa Y., Ito S., Nakamichi N., Mizoguchi T., Niinuma K., Yamashino T., Mizuno T. (2007). Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 48: 925–937. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N., Sun T.P., Gubler F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14 (Suppl): S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Wang W., Guo X., Yue J., Huang Y., Xu X., Li J., Hou S. (2014). Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet. 10: e1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Foster K.R., Morgan P.W., Chory J. (1996). Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 112: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo N.J., Vriezen W.H., Beemster G.T., Van Der Straeten D. (2003). Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 33: 989–1000. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.-H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229. [DOI] [PubMed] [Google Scholar]
- Su Y., Wang S., Zhang F., Zheng H., Liu Y., Huang T., Ding Y. (2017). Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell 29: 2197–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wang Z., Casas-Mollano J.A., Xu J., Riethoven J.-J.M., Zhang C., Cerutti H. (2015). Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D., Ori N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144: 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C.-K., Chang C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang S., et al. (2015). C-terminal domains of a histone demethylase interact with a pair of transcription factors and mediate specific chromatin association. Cell Discov. pii: 15003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.-L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.-O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P. (2011). Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]