Abstract
Local axonal protein synthesis plays a crucial role in the formation and function of neuronal circuits. Understanding the role of this mechanism in specific circuits requires identifying the protein composition and mRNA cargos of the ribonucleoprotein particles (RNPs) that form the substrate for axonal translation. FXGs (Fragile X granules) are axonal RNPs present in a stereotyped subset of mature axons in the intact brain that contain one or more of the Fragile X related (FXR) proteins (FMRP, FXR2P, and FXR1P) along with mRNA and ribosomes. Here we performed a systematic survey of the FXR protein composition and mRNA association of FXGs in the brain. We have identified four FXG types that can be categorized based on their FXR protein complement. All FXGs contain FXR2P, with FMRP and/or FXR1P present in circuit-selective subsets. Individual neuronal cell types predominantly express a single FXG type, with FMRP-containing FXGs the most prevalent in forebrain neurons. All FXG types associate with ribosomes and mRNA, but the specific mRNA cargos are a function of FXG type, brain region and neuron class. Transcripts for β-catenin and its regulator APC associate with a subset of forebrain FXGs. Moreover, both these transcripts can colocalize within individual FXGs, suggesting that the axonal translation of functionally related proteins may be coordinately regulated with high spatiotemporal resolution. Cell type-dependent expression of specific RNP types with distinct mRNA cargos, such as FXGs, presents a potential mechanism for regulating local translation and its output in a circuit-dependent manner.
Keywords: axonal translation, local protein synthesis, RNA binding proteins, RRID: AB_10805421, RRID: AB_2313703, RRID: AB_2661850, RRID: AB_476964, RRID: AB_528262
1 | INTRODUCTION
The formation and plasticity of neural circuits requires precise spatio-temporal regulation of the neuronal proteome. The importance of this regulation is highlighted by the strong association between dysregulated protein synthesis and cognitive deficits, notably in autism and autism-related disorders such as PTEN hamartoma syndrome, tuberous sclerosis, and Fragile X syndrome (FXS) (Kelleher & Bear, 2008). FXS is the most common single-gene form of intellectual disability and autism and is caused by mutations in the gene encoding FMRP (Fragile X mental retardation protein), an RNA-binding protein that serves as a key regulator of neuronal protein synthesis. Further, common variants in the genes encoding the Fragile X-related (FXR) proteins FMRP, FXR1P, and FXR2P modify autistic traits in the general population (Stepniak et al., 2015). Loss of FMRP leads to profound changes in synaptic function that are thought to underlie the developmental delay, decreased cognitive function, autism, epilepsy, anxiety, and hypersensitivity to sensory stimuli characteristic of this disorder. Notably, while FXS patients exhibit general developmental delay, not all domains are affected equally (Roberts et al., 2009). Moreover, FXS patients show region-selective abnormalities in brain morphology and connectivity (Bruno et al., 2017; Hall, Jiang, Reiss, & Greicius, 2013; Hoeft et al., 2010). These findings suggest that FMRP plays circuit-dependent roles in the regulation of neuronal protein expression.
One mode by which FMRP exerts precise control over the neuronal proteome is by regulating local translation in both the dendritic and axonal arbors. The localization and translation of mRNAs are dependent on their interaction with ribonucleoprotein particles (RNPs), or RNA granules. The brain region-dependent effects seen in FXS patients raise the possibility that both FMRP-containing RNPs and the mRNAs that they regulate may differ among circuits. Elucidating how, when and where FMRP-dependent translational regulation functions in the brain therefore requires determining the protein composition, mRNA cargo and circuit-selective expression of FMRP-containing granules. In axons, FMRP and the other FXR proteins localize to discrete, circuit-selective RNPs termed Fragile X granules (FXGs) (Christie, Akins, Schwob, & Fallon, 2009). FXGs are expressed exclusively in axons that have synaptically integrated into circuits where they associate with ribosomes and mRNA and influence the composition of the axonal proteome (Akins et al., 2017). The circuits that contain FXGs as well as the association of these granules with ribosomes and mRNA are conserved across mice, rats and humans (Akins et al., 2017), suggesting that their study in mice will elucidate axonal translation in the human brain.
Neurons exhibit remarkable circuit-dependent structural and functional diversity in their axonal arbors. Circuit-selective expression of axonal RNPs may be one mechanism supporting this diversity. As a first step to address this question, we asked whether FXGs comprise a family of RNPs that differ among neural circuits in their protein composition and mRNA cargoes. We find that FXGs can be classified into four types based on their FXR protein composition. Strikingly, within brain regions, FXG type was homogeneous: most FXGs in each circuit contain the same combination of Fragile X proteins. Although all four FXG types associate with ribosomes and mRNA, the specific mRNA cargoes varied among circuits. The mRNAs encoding β-catenin and APC (adenomatous polyposis coli) are associated exclusively with forebrain FXGs in select circuits, where both transcripts could be found together within individual FXGs. Together, these findings suggest that FMRP-regulated axonal translation may mediate distinct cellular roles in different neuronal circuits.
2 | MATERIALS AND METHODS
2.1 | Animals
All work with animals was performed in accordance with protocols approved by the Institutional Animal Care and Use Committees of Brown and Drexel Universities. C57Bl/6 wild-type mice or Fmr1 knockout mice on the C57Bl/6 background were deeply anesthetized by isoflurane inhalation before intracardiac perfusion with room temperature HBS (0.1M HEPES, pH 7.4; 150 mM sodium chloride) containing 1 U/mL heparin and 0.1% sodium nitrite followed by perfusion with room temperature PBS (0.1M phosphate, pH 7.4; 150 mM sodium chloride) containing 4% paraformaldehyde. After perfusion, animals were decapitated and intact brains were carefully removed. After washing in PBS, brains were transferred to PBS containing 30% sucrose until the brains sank. Brains were then embedded in OCT medium by rapid freezing and stored at −80°C until sectioning. Free-floating coronal sections of OCT-embedded brains were prepared using a Leica cryostat at 50 μm and either used the same day or stored at −20°C in antifreeze solution (50 mM phosphate pH 7.4, 30% sucrose, 30% ethylene glycol, 1% polyvinyl pyrrolidone) until use for immunolabeling.
2.2 | Immunohistochemistry
Tissue stored in antifreeze was first washed three times in PBS (10 mM phosphate pH 7.4, 150 mM NaCl) before antigen retrieval, while tissue stained immediately after cutting was directly treated for antigen retrieval. To improve antibody accessibility to epitopes, tissue sections were first heated in 0.01M sodium citrate (pH 6.0) for 30 min at 75°C. Tissue was then treated with blocking solution [PBST (10 mM phosphate buffer, pH 7.4, and 0.3% Triton X-100) and 1% blocking reagent (Roche)] for 30 min to block nonspecific binding sites. Sections were then treated with blocking solution plus primary antibody (Table 1) overnight before washing for 5 min with PBST. For secondary detection, tissue was incubated with appropriate secondaries in blocking solution for 1 hr. For all secondary antibodies, each lot was validated to ensure no cross reactivity with inappropriate primary antibodies. Tissue was then washed for 5 min with PBST, mounted in NPG mounting medium (4% n-propylgallate, 60% glycerol, 5 mM phosphate pH 7.4), coverslipped, and sealed with nail polish.
TABLE 1.
Antibodies used in this study
| Antibody name, target, and host | Immunogen and Specificity | Source | Concentration |
|---|---|---|---|
| 1G2 anti FXR2P Mouse IgG1 monoclonal |
Raised against recombinant FXR2 residues 414–658. Recognizes a single band of the appropriate size (Zang et al., 2009). | Developmental Studies Hybridoma Bank (ascites form). RRID: AB_528262 |
1:1000 |
| A42 anti FXR2P Mouse IgG1 monoclonal |
Raised against human FXR2P amino acids 12–426; recognizes a single band of the appropriate size by Western and does not cross react with either FMRP or FXR1P (Zhang et al., 1995). | Sigma Aldrich (Cat # F1554). RRID: AB_476964 |
1:500 |
| 2F5-1 anti FMRP Mouse IgG2b monoclonal |
Raised against amino acids 1–204 of human FMRP. Recognizes bands of the appropriate sizes by Western, does not give signal in either blots or sections derived from Fmr1 null mice (Christie et al., 2009; Gabel et al., 2004). | In house. Concentrated supernatant from hybridoma cells. RRID: AB_10805421 |
5 μg/mL |
| Ab2107 anti FXR1P Rabbit polyclonal |
Raised against amino acids 483–500 of human FXR1P. Recognizes bands of appropriate length by Western, does not cross-react with FMRP or FXR2P (Tamanini et al., 1999). | Gift of Dr. A. Hoogeveen. RRID: AB_2661850 |
1:1000 |
| Y10b anti 5S/5.8S rRNA Mouse IgG2a monoclonal |
Autoimmune antibody from a lupus mouse model that recognizes 5S and 5.8S rRNA (Garden, Canady, Lurie, Bothwell, & Rubel, 1994; Lerner, Lerner, Janeway, & Steitz, 1981). FXG-associated signal is sensitive to RNase treatment (Akins et al., 2017). | In house. Supernatant from hybridoma cells. RRID: AB_2313703 |
1:1000 |
All primary antibodies were detected with secondary antibodies conjugated to Alexa fluor 488, 555 or 647 from Life Technologies used at 1:1000. Isotype-specific secondary antibodies were used to detect the monoclonal primary antibodies.
2.3 | FXG composition quantification
Fluorescent micrographs of brain sections immunostained for FMRP, FXR1P, and FXR2P were collected as above. FXGs were identified based on morphology using FXR2P signal as previously described (Akins et al., 2017; Akins, LeBlanc, Stackpole, Chyung, & Fallon, 2012; Christie et al., 2009). To assess protein composition, each FXR2P-containing FXG was then manually annotated as to whether it also colocalized with FMRP or FXR1P signal. To determine mRNA association, each FXG was manually annotated to determine whether it colocalized with mRNA encoding either β-catenin or APC. To minimize false negatives, only images in which all probes gave clear signal in neuronal somata were analyzed.
2.4 | In situ hybridization
This protocol was performed as previously described (Akins et al., 2017). In brief, fresh frozen brains were sectioned at 20 μm, mounted on slides, and stored at −20°C until use. All subsequent steps were performed at room temperature unless otherwise noted. On the day of staining, slides were allowed to warm to room temperature, fixed in PBS +4% PFA for 10 min, and washed 3× in PBS. They were treated in 0.01M sodium citrate, pH 6 for 30′ at 75°C. They were then treated for 10 min in 0.2M HCl and 2′ in PBS +1% Triton X-100. They were subsequently rinsed twice for 1 min each in PBS before a 10-min equilibration in 2× SSC +10% formamide. For hybridization, sections were incubated overnight at 37°C in hybridization solution containing Stellaris fluorophore-labeled oligonucleotide probes (Biosearch Technologies; used at 250 nM; sequences in Table S1, Supporting Information). Probe sets were labeled with either Quasar 570 or Quasar 670. For detection of FXGs, the A42 monoclonal against FXR2P was used along with a secondary antibody conjugated to Alexa 488. Sections were then rinsed two times for 30′ each at 37°C in 2× SSC +10% formamide. Sections were subsequently rinsed in 2× SSC followed by PBST. They were then incubated in blocking solution plus a secondary antibody for 1 hr to detect A42. Sections were rinsed 3× in PBST, mounted in NPG mounting medium, and then imaged. The fidelity of the Ctnnb1 and Apc signal was confirmed as two probe sets that recognize nonoverlapping portions of the transcript (Table S1, Supporting Information) gave identical signal. The specificity of the oligo(dT) probe was confirmed by substituting an oligo(dA) probe, which produced no detectable signal in brain sections (Akins et al., 2017).
2.5 | Imaging
Images were collected using a Zeiss LSM 510 confocal microscope. To limit the potential detection of inappropriate “bleed through” signal, double- and triple-labeled images were collected in two separate imaging tracks. In the first track, the green signal (Alexa 488, excited at 488 nm and with signal detected from 500 to 545 nm) was detected along with the far-red signal (Alexa 647 or Quasar 670, excited at 633 nm with emission detected from 649 to 713 nm) using simultaneous excitation and detection by two photomultiplier tubes. In the second track, the red (Alexa 555 or Quasar 570) signal was excited at 561 nm and the emission from 575 to 615 nm was recorded using a single photomultiplier tube.
3 | RESULTS
3.1 | Four classes of FXGs in axons of intact brain
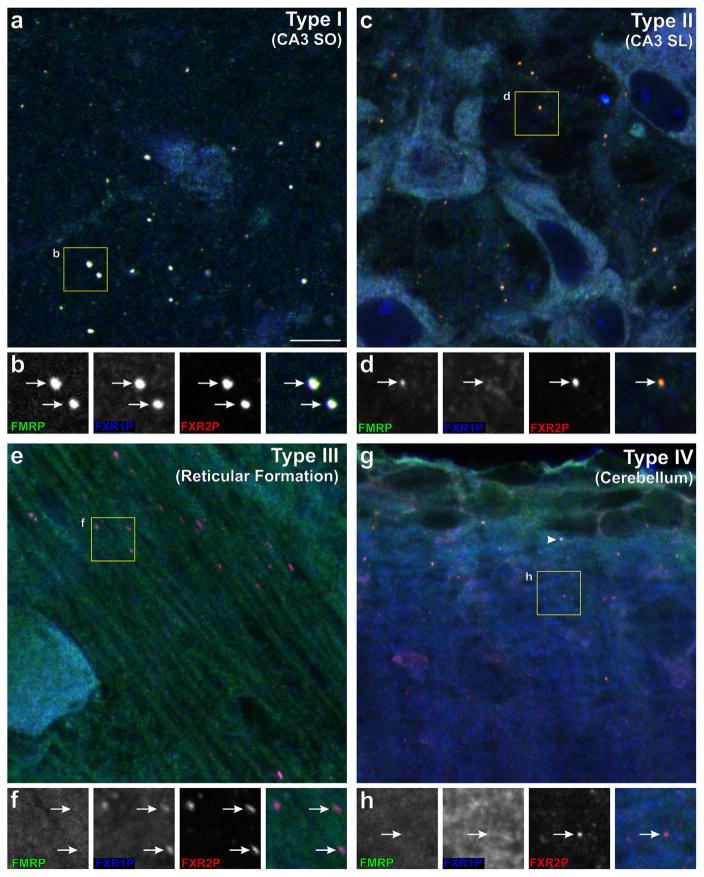
In previous studies, we showed that FXGs can be identified in circuit-selective axonal populations based on their distinctive morphology and localization in micrographs of FXR protein immunostaining (Akins et al., 2017, 2012; Christie et al., 2009). The assignment of FXGs to axons was confirmed by a range of methods including immunoelectron microscopy, ablation and genetic experiments (Akins et al., 2017, 2012; Christie et al., 2009). Here we asked whether FXGs comprise a family of FXR protein-containing RNPs with discrete protein compositions. We immunostained brain sections simultaneously for all three FXR proteins at P15 (the age at which FXGs are most broadly expressed in the mouse brain)(Akins et al., 2017; Christie et al., 2009). We then identified FXGs, all of which contain FXR2P, across the brain and asked whether they also contained FMRP and/or FXR1P. This analysis revealed that FXGs can be classified into four types (I – IV) based on their FXR protein complement: Type I: FXR2P, FMRP, FXR1P; Type II: FXR2P, FMRP; Type III: FXR2P, FXR1P; Type IV: FXR2P only (Figure 1, Table 2). As described below, FXG composition is circuit-dependent with an individual FXG type predominant in a given brain region (Tables 3 and 4).
FIGURE 1.
Four distinct types of FXGs show region-specific expression in the brain axons. Sections of P15 mouse brain were stained for FMRP (green), FXR2P (red), and FXR1P (blue). (a, b) Type I FXGs (containing FXR2P, FXR1P and FMRP, arrows in B) in stratum oriens of hippocampal area CA3. (c, d) Type II FXGs (arrows in D) in stratum lucidum of hippocampal area CA3 contain FMRP and FXR2P but not FXR1P. (e, f) Type III FXGs in reticular formation axons in the medulla contain FXR2P and FXR1P, but not FMRP (arrows in F). (g, h) Type IV FXGs in the deep cerebellar molecular layer contain FXR2P but neither FMRP nor FXR1P (arrows in H). Type I FXGs in parallel fibers in the superficial molecular layer (containing all three FXR proteins; arrowhead in G). Scale bar = 7.5 μm in A,C,E,G; 4 μm in B,D,F,H. See also Tables 3 and 4
TABLE 2.
FXR protein composition of the four FXG types
| Type | FXR2P | FMRP | FXR1P |
|---|---|---|---|
| I | + | + | + |
| II | + | + | − |
| III | + | − | + |
| IV | + | − | − |
Presence of an FXR protein in a granule type is indicated by a “+”.
TABLE 3.
FXG types in individual brain regions
| Brain Region | Type I | Type II | Type III | Type IV |
|---|---|---|---|---|
| Forebrain | ||||
| Olfactory Bulb Glomeruli | 100% (421/421) | |||
| Thalamus | 100% (114/114) | |||
| Striatum | 100% (47/47) | |||
| Neocortex/Subcortical White Matter | 99.2% (237/239) | 0.8% (2/239) | ||
| Olfactory Limb of the Anterior Commissure | 80.1% (141/176) | 19.9% (35/176) | ||
| Hippocampus CA3 Stratum Oriens | 91.4% (53/58) | 6.9% (4/58) | 1.7% (1/58) | |
| Hippocampus CA3 Stratum Lucidum | 1.9% (2/103) | 98.1% (101/103) | ||
| Midbrain/Hindbrain | ||||
| Midbrain/Pons | 1.4% (3/220) | 71.4% (157/220) | 27.3% (60/220) | |
| Superficial Cerebellar Molecular Layer | 100% (13/13) | |||
| Deep Cerebellar Molecular Layer/Granule Cell Layer | 10.5% (2/19) | 5.3% (1/19) | 84.2% (16/19) | |
| Rostral Medulla | 3.7% (2/54) | 1.9% (1/54) | 53.7% (29/54) | 40.7% (22/54) |
| Caudal Medulla | 9.6% (19/197) | 64.5% (127/197) | 25.9% (51/197) | |
FXGs of each type observed in selected brain regions. Numbers are depicted as percent of the total observations with absolute numbers in parentheses. Data represent summed observations from 4 animals. FXG types that comprise more than 40% of the total FXGs in each brain region are indicated in bold.
TABLE 4.
Predominant FXG type in individual circuits
| FXG Type | Circuits | FXG-associated transcripts |
|---|---|---|
| Type I (FXR2P, FMRP, FXR1P) | Hippocampal associational fibers | APC, β-catenin |
| Thalamocortical axons | APC | |
| Corticocortical axons | APC, β-catenin | |
| Cerebellar parallel fibers | ||
| Olfactory limb of the anterior commissure (non-crossing fibers) | ||
|
| ||
| Type II (FXR2P, FMRP) | Hippocampal mossy fibers | APC, MAP1B |
| Olfactory nerve | OMP, NCAM | |
|
| ||
| Type III (FXR2P, FXR1P) | Reticular formation | |
|
| ||
| Type IV (FXR2P) | Deep cerebellar molecular layer | |
Indicated are circuits in which each FXG type predominates. Specific transcripts associated with FXGs in each circuit are indicated (see also Akins et al., 2017).
3.2 | FXGs in the forebrain
Almost all of the forebrain FXGs identified in the current study contained FMRP (Table 3). Quantification revealed that single FXG types predominated in given brain regions. Type I FXGs were the most common class in the forebrain (Tables 3 and 4) and were present in thalamocortical and corticocortical axons in the thalamus (100%), striatum (100%) and neocortex (99%). Type I FXGs also predominated in the olfactory limb of the anterior commissure (80%) and the stratum oriens of hippocampal area CA3 (91%) (Figure 1a–b, Tables 3 and 4). Type II FXGs predominated in two forebrain circuits: mossy fibers in stratum lucidum of hippocampal area CA3 (98%) and olfactory sensory neuron axons in the olfactory bulb (100%; Figure 1c–d, Tables 3 and 4). Olfactory sensory neuron axons are the only axonal population to contain substantial numbers of FXGs into adulthood (Akins et al., 2017; Christie et al., 2009). FXGs in olfactory sensory neuron axons of 3- to 5-month-old animals were also Type II (data not shown), suggesting that FXG composition within this brain region is stable across the lifespan. Type II granules were also found in the olfactory limb of the anterior commissure, where they made up 20% of the total FXGs (Table 3). The homogeneity of FXG composition in these well-defined circuits suggests that individual forebrain neuronal types contain predominantly a single class of FXGs.
3.3 | FXGs in the midbrain and hindbrain
We next examined FXG composition within circuits of the midbrain and hindbrain. In contrast to the observations in forebrain, FMRP was not a component of most midbrain and hindbrain FXGs (Tables 3 and 4). The one FMRP-containing FXG population in the hindbrain comprised Type I FXGs in the superficial molecular layer of the cerebellar cortex (Figure 1g), where FXGs are found in parallel fibers (Christie et al., 2009). Deeper within the cerebellar molecular layer was a second population of FXGs that were 84% Type IV (Figure 1g–h). Midbrain, pons, and medulla contained a mixture of all four FXG types. Type III granules were the most common and comprised nearly all of the FXGs in the reticular formation (Figure 1e–f). Thus, FXG composition was more heterogeneous in the midbrain and hindbrain than in the forebrain, potentially reflecting the diversity of axon populations found in these regions.
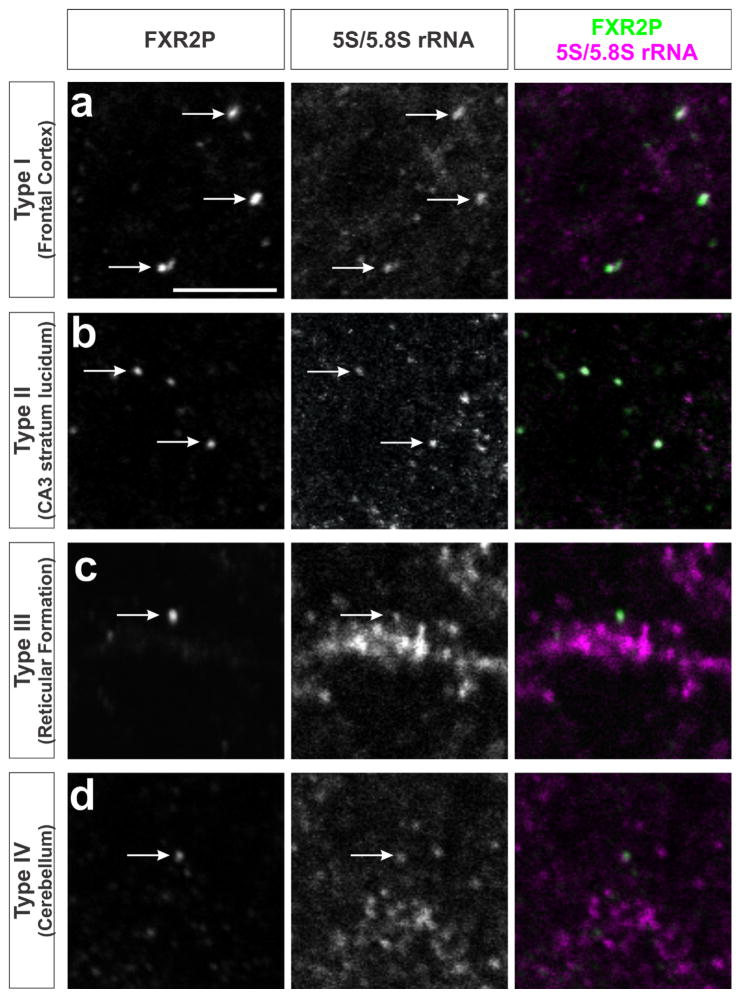
3.4 | All FXG types associate with ribosomes
Previous studies have shown that FXGs in the forebrain associate with translational machinery including ribosomes (Akins et al., 2017). Here we investigated whether ribosome association is a general feature of all FXG types. We performed immunohistochemistry using an antibody for FXR2P together with an antibody (Y10b) that recognizes 5S and 5.8S ribosomal RNA (rRNA; see Methods). Figure 2 shows that ribosomes colocalized with FXGs throughout the brain, including with granules in motor cortex (Type I), hippocampal mossy fibers (Type II), reticular formation (Type III) and the deep cerebellar molecular layer (Type IV). Thus, all four types of FXGs associate with ribosomes.
FIGURE 2.
Ribosomes are associated with all four FXG types. Confocal micrographs of co-immunostaining for FXR2P to identify FXGs (green) and Y10b antibody to identify 5S and 5.8S rRNA (red) in different brain regions. Ribosome-associated FXGs (arrows) include: (a) Type I FXGs in frontal cortex; (b) Type II FXGs in hippocampal mossy fibers; (c) Type III FXGs in the reticular formation within the medulla; and (d) Type IV FXGs in the deep cerebellar molecular layer. Scale bar = 5 μm
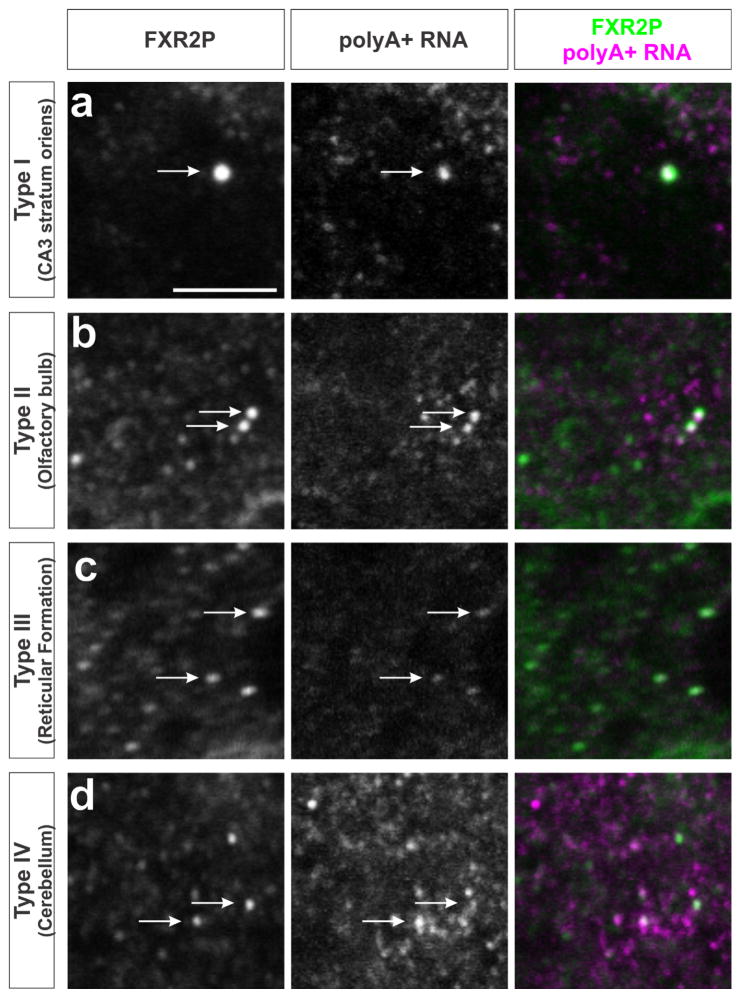
3.5 | All FXG types associate with polyA+ RNA
In previous work, we showed that polyA+ RNA is associated with FXGs in the forebrain (Akins et al., 2017). Here we extended this analysis and asked whether RNA associates with all types of FXGs throughout the brain. In situ hybridization using an oligo(dT) probe to detect polyA+ RNA combined with FXR2P immunostaining revealed that polyA+ signal was detected in FXGs throughout the brain (Figure 3). RNA associated with FXGs in stratum oriens of hippocampal area CA3 (Type I), olfactory glomeruli (Type II), reticular formation (Type III) and the deep cerebellar molecular layer (Type IV). PolyA+ RNA therefore associates with all four types of FXGs.
FIGURE 3.
All four FXG types associate with polyA+ RNA. Confocal micrographs of immunostaining for FXR2P (green) to identify FXGs, and in situ hybridization for polyA to identify polyadenylated RNAs (red). PolyA-associated FXGs (arrows) include: (a) Type I FXGs in CA3 associational fibers; (b) Type II FXGs in olfactory sensory neuron axons; (c) Type III FXGs in the reticular formation within the medulla; and (d) Type IV FXGs in the deep cerebellar molecular layer. Scale bar = 5 μm
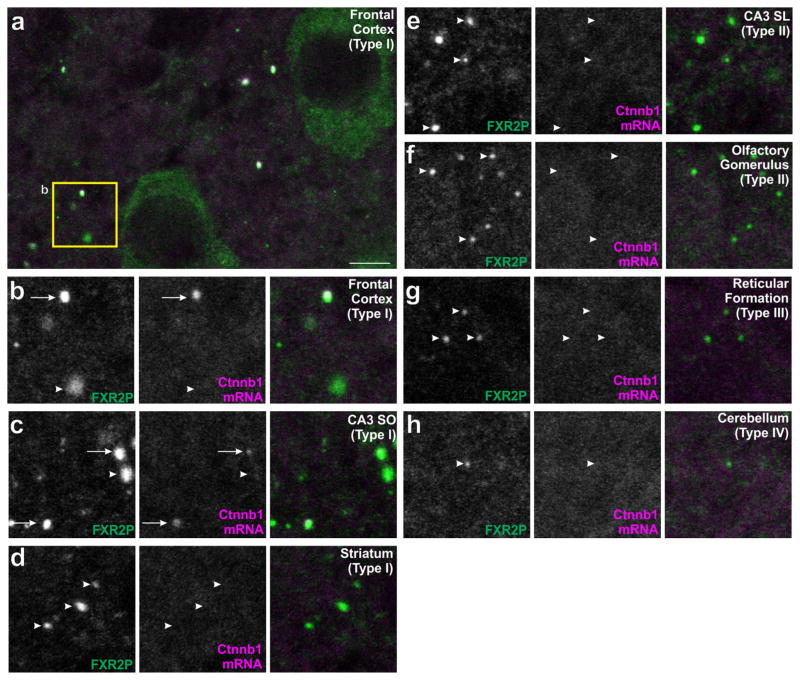
3.6 | Circuit-selective subpopulations of FXGs contain the Ctnnb1 mRNA
We next asked whether individual FXG types selectively associate with specific mRNA cargos. We previously identified Ctnnb1 mRNA (encoding β-catenin) as an FXG cargo in cortical axons (Akins et al., 2017). Figure 4a–c shows that Ctnnb1 mRNA is associated with Type I FXGs in hippocampal CA3 associational fibers as well as in neocortical axons. In contrast, Ctnnb1 mRNA did not associate with Type I FXGs in other brain regions, including those in thalamocortical axons in the striatum (Figure 4d). Moreover, Ctnnb1 mRNA does not associate with Type II, III, or IV FXGs (as seen in hippocampal mossy fibers, olfactory sensory neuron axons, axons of the reticular formation and axons in the deep cerebellar cortex; Figure 4e–h, Table 3). Thus β-catenin mRNA is a component of a circuit-selective subset of Type I FXGs.
FIGURE 4.
β-catenin mRNA associates with a region-dependent subset of Type I FXGs. Brain sections were double-labeled using immunostaining for FXR2P (green) to identify FXGs and in situ hybridization to identify the β-catenin mRNA (Ctnnb1; red). (a–b) In frontal cortex, the Ctnnb1 mRNA associated with a subset of Type I FXGs but was absent from other FXGs. (c) FXGs in associational fibers in CA3 stratum oriens contain the Ctnnb1 transcript. In contrast, Ctnnb1 mRNA did not colocalize with Type I FXGs in (d) thalamocortical axons in the striatum. Ctnnb1 mRNA was not found in Type II, III, or IV FXGs including those in (e) hippocampal mossy fibers in CA3 stratum lucidum, (f) olfactory sensory neuron axons in olfactory glomeruli, (g) axons in the reticular formation, or (h) axons in the deep cerebellar molecular layer. Arrows indicate FXGs that colocalize with Ctnnb1. Arrowheads indicate FXGs that do not colocalize with Ctnnb1. Scale bar = 10 μm in A, 5 μm in B–H
3.7 | Circuit-selective subpopulations of FXGs contain the Apc mRNA
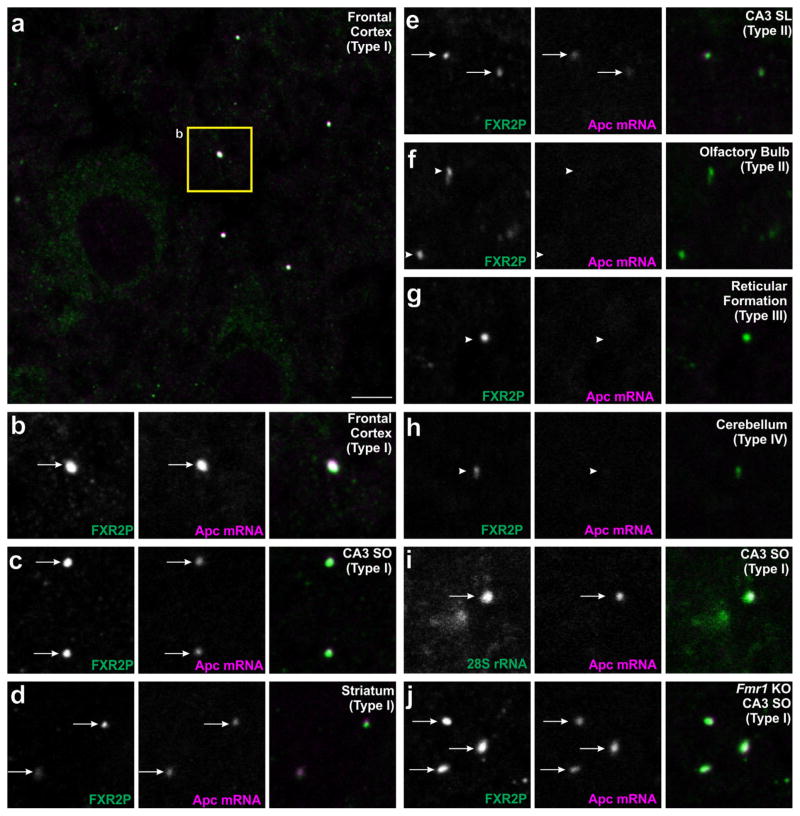
We next asked whether additional mRNAs also show circuit-selective FXG association. Another candidate for FXG association is the Apc mRNA, which encodes adenomatous polyposis coli, a negative regulator of β-catenin protein levels (Stamos & Weis, 2013). Apc mRNA directly binds to FMRP and localizes to axons of cultured cortical neurons (Darnell et al., 2011; Taylor et al., 2009). In situ hybridization for Apc combined with FXR2P immunolabeling revealed that Apc mRNA is associated with FXGs in several circuits (Table 3). Apc mRNA associated with Type I FXGs in both motor cortex (Figure 5a–b) and striatum (Figure 5d). We also found Apc-containing FXGs in hippocampal circuits including both Type I in CA3 stratum oriens (Figure 5c) as well as Type II in CA3 stratum lucidum (Figure 5e). In contrast, we did not observe Apc mRNA in FXGs in olfactory sensory neuron axons (Type II, Figure 5f), axons of the reticular formation (Type III, Figure 5g), or the deep cerebellar cortex (Type IV, Figure 5h). Finally, Apc mRNA and ribosomes are colocalized in individual FXGs (Figure 5j). The Apc mRNA is thus a component of region-selective subsets of Type I and Type II FXGs within select forebrain circuits.
FIGURE 5.
APC mRNA associates with a region-dependent subset of FXGs. Brain sections were double-labeled by in situ hybridization for Apc (red) along with either immunostaining for FXR2P (green; A–H,J) or in situ hybridization for 28S rRNA (green; I). Apc-containing FXGs were observed in a region-dependent subset of Type I FXGs including those in (a–b) axons in frontal cortex, (c) associational fibers in CA3 stratum oriens, and (d) thalamocortical axons in striatum. Apc mRNA also associated with Type II FXGs in (e) hippocampal mossy fibers but not Type II FXGs in (f) olfactory sensory neuron axons in the olfactory bulb. Apc mRNA was not observed in Type III or Type IV FXGs including those in (g) axons in the reticular formation in the medulla or (h) axons in the deep cerebellar molecular layer. (i) Double in situ hybridization for Apc and 28S rRNA showed that Apc-associated FXGs colocalize with ribosomes in CA3 associational fibers. (j) FXGs associate with Apc in CA3 associational fibers in an Fmr1 null mouse. Arrows indicate FXGs that colocalize with Apc. Arrowheads indicate FXGs that do not colocalize with Apc. Scale bar = 10 μm in A, 5 μm in B–L
The results described above show that Apc mRNA is only found in FXG populations that contain FMRP (Type I and II; Table 3). We therefore used Fmr1 null mice to test whether FMRP is required for the association of Apc mRNA with FXGs. In situ hybridization for Apc combined with immunofluorescence for FXR2P revealed that, as in wild type mice, Apc mRNA associated with FXGs in both cortex and hippocampus of Fmr1 null animals (Figure 5i). These findings are consistent with previous observations that FXG association with both total polyA+ RNA and Ctnnb1 transcript is equivalent in wild type and Fmr1 mice (Akins et al., 2017). Thus FMRP confers neither FXG Type- nor circuit-specificity of Apc mRNA association with FXGs.
3.8 | Ctnnb1 and Apc mRNAs colocalize in individual FXGs
As Apc and Ctnnb1 mRNAs were both found in FXGs within the same circuits, we asked whether these two RNAs might define distinct nonoverlapping populations of FXGs or whether these mRNAs instead coexist within individual FXGs. Double in situ hybridization for Apc and Ctnnb1 mRNAs combined with FXR2P immunofluorescence showed that both transcripts colocalized within individual FXGs in cortical and hippocampal circuits (Figure 6). Both mRNAs were readily detected in FXGs in frontal cortex. We therefore quantified the colocalization of these mRNAs in FXGs in this region (n = 3 images from 3 animals). In frontal cortex, 58.7% (61/104) of FXGs contained both Apc and Ctnnb1 mRNAs, 8.7% (9/104) contained Apc alone, 7.7% (8/104) contained Ctnnb1 alone, and 25% (26/104) contained neither of these transcripts. Individual FXGs can therefore contain multiple transcripts that encode proteins with antagonistic functions in the same pathway.
FIGURE 6.
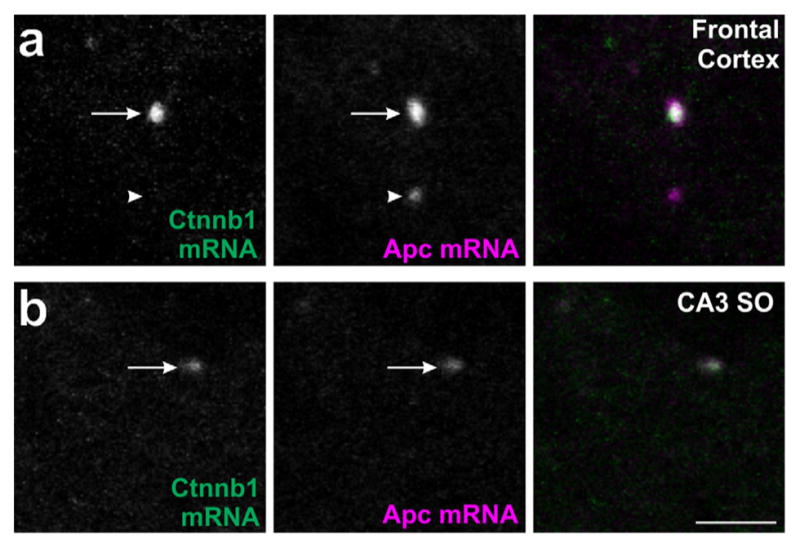
APC and β-catenin mRNAs colocalize to individual FXGs. Confocal micrographs of mouse brain sections double-labeled by in situ hybridization for Apc (red) and Ctnnb1 (green) mRNAs. Individual FXGs contain both the Apc and Ctnnb1 mRNAs in axons in (a) frontal cortex and (b) CA3 stratum oriens. Arrows indicate FXGs that colocalize with Apc. Arrowheads indicate FXGs that do not colocalize with Apc. Scale bar = 5 μm
4 | DISCUSSION
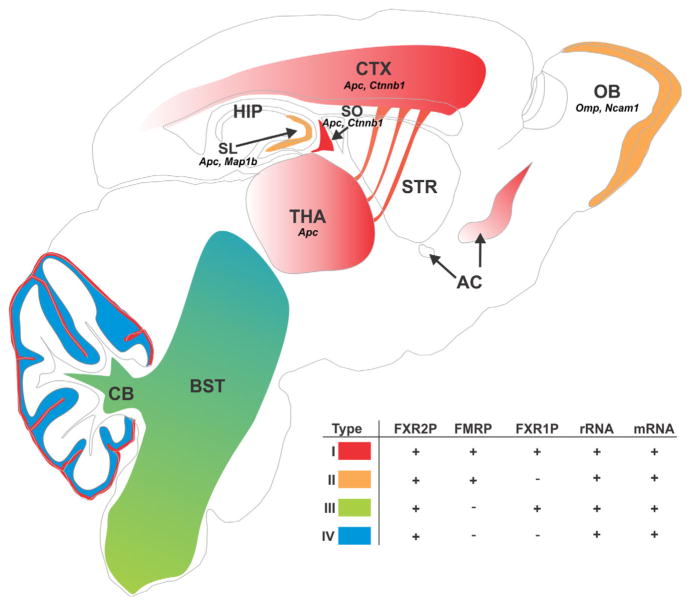
In this study, we investigated FXG composition as a means to elucidate the roles of these axonal RNPs in diverse brain circuits. The systematic analysis of FXG composition and distribution presented here revealed that FXGs comprise a family of related FXR protein-containing granules that are present in axons in restricted circuits throughout the brain. FXGs can be categorized into four granule types based on their FXR protein composition. Strikingly, FXG composition is a function of both brain region and individual neuronal type, with single FXG types predominating in most circuits (Tables 3, 4; Figure 7). FMRP-containing Type I and Type II FXGs are the major forebrain granules, while Type III and Type IV FXGs (which lack FMRP) are most prevalent in the midbrain and hindbrain. Ribosomes and mRNA associate with all four FXGs, but the mRNA cargos identified to date are strikingly circuit-specific. Moreover, individual FXGs can simultaneously associate with multiple transcripts, suggesting that axonal translation could coordinate the local synthesis of proteins with related functions. Finally, these findings suggest that RNP diversity may be a more general mechanism for circuit- and cell type-dependent regulation of neuronal protein synthesis in both the axonal and somatodendritic compartments.
FIGURE 7.
Distribution and composition of FXG types across the brain. All FXGs contain FXR2P and are in axons. FXGs can be subdivided into four types based on whether they contain FMRP and/or FXR1P. All four FXG types associate with ribosomes and mRNA. Identified FXG-associated mRNAs are indicated (Apc, Ctnnb1, Map1b, Ncam1, Omp) (see also Akins et al., 2017). Type I FXGs (red) predominate in the forebrain, where they are most abundant in motor circuits, and are also present in cerebellar parallel fibers. Type II FXGs (orange) comprise >98% of the granules in olfactory sensory neuron axons and hippocampal mossy fibers. Type III FXGs (green) predominate in the brainstem. Type IV FXGs (blue) are seen in the deeper areas of the cerebellar molecular layer and are also present in rostral brainstem circuits. AC: anterior commissure; BST: brain stem; CB: cerebellum; CTX: neocortex; HIP: hippocampus; OB: olfactory bulb; SL: stratum lucidum of hippocampal area CA3; SO: stratum oriens of hippocampal area CA3; STR: striatum; THA: thalamus. See also Tables 3 and 4
4.1 | Potential roles for FXR proteins in FXGs
FXR2P is present in all FXG types and is critical for their expression (Table 2) (Christie et al., 2009). This unique role for FXR2P among the FXR proteins could be mediated at least in part by two of its distinctive structural properties. First, FXR2P is the only FXR protein that is N-terminally myristoylated (Stackpole, Akins, & Fallon, 2014), a posttranslational modification that is important for protein-lipid and protein-protein interactions (Resh, 2016; Taniguchi, 1999). Mutant FXR2P that cannot be myristoylated is abnormally distributed within the axonal arbor of cultured neurons (Stackpole et al., 2014). Second, FXR2P is distinctive among the FXR proteins in its low complexity domain, a class of domains known to play critical roles in the formation of a wide range of RNA granules through protein-protein and protein-RNA interactions (Calabretta & Richard, 2015). For example, FXR2P and FMRP are 93% similar in their Agenet domains but only 36% similar in their low complexity domains. Taken together, these observations suggest that myristoylation-mediated association with cell membranes as well as low complexity domain-dependent interactions may be important for FXR2P-directed FXG localization within axonal arbors in the brain.
The differential inclusion of FMRP and/or FXR1P in FXGs may influence how these granule types regulate axonal protein expression. Studies in model systems have shown that FXR proteins can have distinct functions. For example, FMRP, but not FXR1P or FXR2P, represses translation in several non-neuronal systems (Darnell et al., 2011; Laggerbauer, Ostareck, Keidel, Ostareck-Lederer, & Fischer, 2001). In neurons, both FMRP and FXR1P negatively regulate translation of their associated mRNAs, while FXR2P positively regulates translation (Cook et al., 2014; Fernández et al., 2015; Guo et al., 2015). In addition, specific FMRP splice forms, which can also be circuit-selective, differentially affect axonal arbor morphology (Brackett et al., 2013; Zimmer, Doll, Garcia, & Akins, 2016). Finally, FXR proteins can also influence local protein expression via posttranslational mechanisms. For example, FMRP modulates the plasma membrane delivery of both the AMPA receptor subunit GluA2 as well as the calcium channel CaV2.2 independent of translational regulation (Ferron, Nieto-Rostro, Cassidy, & Dolphin, 2014; Guo et al., 2015). FXG composition may thus influence whether FXGs promote or suppress local translation as well as the posttranslational fate of proteins synthesized in circuit-selective axonal compartments.
4.2 | Circuit-dependent FXG association with mRNAs
Our findings suggest that FXGs support circuit-specific translation of specific axonal mRNAs. Individual neuronal cell types express predominantly, if not exclusively, a single FXG type that associates with a stereotypical subset of mRNAs. The FXR protein makeup of these types may influence the cell type-dependent mRNA association with FXGs since both the Ctnnb1 and Apc transcripts are only found in FMRP-containing FXGs. Consistent with this model, the affinity of FMRP-containing complexes for particular mRNA sequences depends on whether FXR1P is also present in that complex (Bechara et al., 2007). On the other hand, our findings show that neither FMRP nor FXR1P is necessary for FXG association with ribosomes or mRNA (Figures 2–3, and 5) (Akins et al., 2017). Further, FXGs themselves are not required for the axonal localization of their associated mRNAs (Akins et al., 2017). Moreover, FMRP-containing FXGs associate with only a characteristic subset of FMRP target mRNAs, and the absence of FMRP in Fmr1 null mice does not detectably alter the circuit-selective FXG-association of these transcripts (Figure 5) (Akins et al., 2017). Interestingly, the absence of FMRP also does not otherwise alter the FXR protein composition of FXGs, since in Fmr1 null mice FXGs that are normally Type I contain FXR1P while those that are normally Type II do not (Christie et al., 2009). Together, these findings indicate that the axonal transport of FXG target mRNAs is likely to be independent of the assembly and transport of the FXGs. Nonetheless, axonal transport of FXGs and their target mRNAs is likely to be coordinately regulated in a neuronal cell type-dependent manner (Korsak, Shepard, & Akins, 2017). Cell type-dependent expression of these targeting mechanisms likely regulates the axonal transport of specific mRNAs, giving rise to the diversity of the axonal transcriptome reported here and in other studies (Baleriola et al., 2014; Gumy et al., 2011; Shigeoka et al., 2016; Taylor et al., 2009; Willis et al., 2005). Neurons thus exhibit diversity in both the transcripts that are translated in axons as well as the mechanisms that regulate their local translation.
4.3 | Potential functions of FXG-regulated axonally synthesized proteins
Our identification of the circuit selectivity of FXG types and their associated mRNAs was based on an analysis of a small number of protein components and mRNA cargos. A full understanding of the impact of FXGs on brain function will require the use of neuronal type-specific tools to identify the full panoply of FXG protein components and mRNA cargos and how these differ among circuits and across development. However, our results suggest that one mechanism whereby FXG-regulated axonal translation could impact presynaptic physiology is by regulating the synaptic vesicle pool, in particular through the modulation of β-catenin levels. β-Catenin is locally translated during presynaptic differentiation and constitutively present at mature presynaptic sites, where it restrains vesicles at synapses and regulates the size of the reserve pool (Bamji et al., 2003; Bamji, Rico, Kimes, & Reichardt, 2006; Taylor, Wu, Tai, & Schuman, 2013). APC negatively regulates β-catenin levels in a Wnt-dependent manner (Stamos & Weis, 2013). Thus, FXG-regulated local synthesis of β-catenin and/or APC, potentially modulated by coincident Wnt signaling, provides an attractive mechanism to link neuronal activity to changes in the synaptic vesicle pool. Notably, the specific effects in a given axon would depend on whether the combination of local protein synthesis and Wnt signaling favored an increase or a decrease in β-catenin levels. For example, frontal cortex contains populations of axons harboring FXGs that associate with only β-catenin, only APC or both transcripts (Figure 6). FXG-regulated local translation in frontal cortex could thus result in either increased or decreased β-catenin levels – with resultant opposite effects on the structure and dynamics of the vesicle pool –depending both on the cell type in which the translation occurs and whether or not a coincident Wnt signal is present. In this way, presynaptic translation may favor strengthening of synapses in some axons and weakening of synapses in others within an individual brain region.
4.4 | FXGs and Fragile X syndrome
Only FXGs in forebrain and cerebellar axons contain FMRP, suggesting that loss of presynaptic translational regulation in Fragile X syndrome would specifically affect synapses in those circuits. Further, since FXG-associated transcripts vary among neuronal types, each FXG-containing circuit is likely affected in distinct fashions in FXS. Notably, loss of FMRP leads to increased expression of its protein targets in FXG-containing axons (Akins et al., 2017). Thus, loss of FMRP in FXS could result in abnormal FXG-regulated axonal translation, leading to circuit-selective alterations in the dosage of presynaptic FMRP targets. Since FXGs are present in the adult human brain throughout life where they associate with ribosomes and mRNA (Akins et al., 2017), changes in presynaptic translation could selectively affect cognitive functions mediated by the forebrain (and perhaps cerebellar cortex), while sparing FXG-related functions elsewhere in the brain. Together, these findings reveal the complexity of axonal RNPs in the mammalian brain and suggest that circuit-dependent alterations in their function contribute to the symptoms of FXS and potentially other neurodevelopmental and neurodegenerative disorders.
Supplementary Material
Acknowledgments
Funding information
National Institute of Mental Health, MH090237;Eunice Kennedy Shriver National Institute of Child Health and Human Development, HD052083
We thank A. Hoogeveen for the gift of the FXR1P antibody. We are grateful to B. McKechnie, C. Schmiedel, and K. Shepard for technical assistance. This work was supported by National Institutes of Health grants HD052083 to JRF and MH090237 to MRA.
Footnotes
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- Akins MR, Berk-Rauch HE, Kwan KY, Mitchell ME, Shepard KA, Korsak LIT, … Fallon JR. Axonal ribosomes and mRNAs associate with fragile X granules in adult rodent and human brains. Human Molecular Genetics. 2017;26(1):192–209. doi: 10.1093/hmg/ddw381. https://doi.org/10.1093/hmg/ddw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins MR, LeBlanc HF, Stackpole EE, Chyung E, Fallon JR. Systematic mapping of fragile X granules in the mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. The Journal of Comparative Neurology. 2012;520(16):3687–3706. doi: 10.1002/cne.23123. https://doi.org/10.1002/cne.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell. 2014;158(5):1159–1172. doi: 10.1016/j.cell.2014.07.001. https://doi.org/10.1016/j.cell.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamji SX, Rico B, Kimes N, Reichardt LF. BDNF mobilizes synaptic vesicles and enhances synapse formation by disrupting cadherin–β-catenin interactions. The Journal of Cell Biology. 2006;174(2):289–299. doi: 10.1083/jcb.200601087. https://doi.org/10.1083/jcb.200601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamji SX, Shimazu K, Kimes N, Huelsken J, Birchmeier W, Lu B, Reichardt LF. Role of β-catenin in synaptic vesicle localization and presynaptic assembly. Neuron. 2003;40(4):719–731. doi: 10.1016/s0896-6273(03)00718-9. https://doi.org/10.1016/S0896-6273(03)00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara E, Davidovic L, Melko M, Bensaid M, Tremblay S, Grosgeorge J, … Bardoni B. Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure. Nucleic Acids Research. 2007;35(1):299–306. doi: 10.1093/nar/gkl1021. https://doi.org/10.1093/nar/gkl1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackett DM, Qing F, Amieux PS, Sellers DL, Horner PJ, Morris DR. Fmr1 transcript isoforms: Association with polyribosomes; regional and developmental expression in mouse brain. PLoS ONE. 2013;8(3):e58296. doi: 10.1371/journal.pone.0058296. https://doi.org/10.1371/journal.pone.0058296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno JL, Hosseini SMH, Saggar M, Quintin EM, Raman MM, Reiss AL. Altered brain network segregation in fragile X syndrome revealed by structural connectomics. Cerebral Cortex. 2017;27(3):2249–2259. doi: 10.1093/cercor/bhw055. https://doi.org/10.1093/cercor/bhw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta S, Richard S. Emerging roles of disordered sequences in RNA-binding proteins. Trends in Biochemical Sciences. 2015;40(11):662–672. doi: 10.1016/j.tibs.2015.08.012. https://doi.org/10.1016/j.tibs.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: A presynaptic fragile X granule expressed in a subset of developing brain circuits. The Journal of Neuroscience. 2009;29(5):1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. https://doi.org/10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Nuro E, Jones EV, Altimimi HF, Farmer WT, Gandin V, … Murai KK. FXR1P limits long-term memory, long-lasting synaptic potentiation, and de novo GluA2 translation. Cell Reports. 2014;9(4):1402–1416. doi: 10.1016/j.celrep.2014.10.028. https://doi.org/10.1016/j.celrep.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, … Darnell RB. FMRP stalls ribosomal translocation on mrnas linked to synaptic function and Autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. https://doi.org/10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E, Li KW, Rajan N, Rubeis SD, Fiers M, Smit AB, … Bagni C. FXR2P exerts a positive translational control and is required for the activity-dependent increase of PSD95 expression. The Journal of Neuroscience. 2015;35(25):9402–9408. doi: 10.1523/JNEUROSCI.4800-14.2015. https://doi.org/10.1523/JNEUROSCI.4800-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron L, Nieto-Rostro M, Cassidy JS, Dolphin AC. Fragile X mental retardation protein controls synaptic vesicle exocytosis by modulating N-type calcium channel density. Nature Communications. 2014;5:3628. doi: 10.1038/ncomms4628. https://doi.org/10.1038/ncomms4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, Fallon JR. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. The Journal of Neuroscience. 2004;24(47):10579–10583. doi: 10.1523/JNEUROSCI.2185-04.2004. https://doi.org/10.1523/JNEUROSCI.2185-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA, Canady KS, Lurie DI, Bothwell M, Rubel EW. A biphasic change in ribosomal conformation during transneuronal degeneration is altered by inhibition of mitochondrial, but not cytoplasmic protein synthesis. The Journal of Neuroscience. 1994;14(4):1994–2008. doi: 10.1523/JNEUROSCI.14-04-01994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GSH, Tung YCL, Zivraj KH, Willis D, Coppola G, … Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98. doi: 10.1261/rna.2386111. https://doi.org/10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Polich ED, Su J, Gao Y, Christopher DM, Allan AM, … Zhao X. Fragile X proteins FMRP and FXR2P control synaptic GluA1 expression and neuronal maturation via distinct mechanisms. Cell Reports. 2015;11(10):1651–1666. doi: 10.1016/j.celrep.2015.05.013. https://doi.org/10.1016/j.celrep.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SS, Jiang H, Reiss AL, Greicius MD. Identifying large-scale brain networks in fragile X syndrome. JAMA Psychiatry. 2013;70(11):1215–1223. doi: 10.1001/jamapsychiatry.2013.247. https://doi.org/10.1001/jamapsychiatry.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, Carter JC, Lightbody AA, Cody Hazlett H, Piven J, Reiss AL. Region-specific alterations in brain development in one- to three-year-old boys with fragile X syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(20):9335–9339. doi: 10.1073/pnas.1002762107. https://doi.org/10.1073/pnas.1002762107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, Bear MF. The Autistic neuron: Troubled translation? Cell. 2008;135(3):401–406. doi: 10.1016/j.cell.2008.10.017. https://doi.org/10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Korsak LIT, Shepard KA, Akins MR. Cell type-dependent axonal localization of translational regulators and mRNA in mouse peripheral olfactory neurons. The Journal of Comparative Neurology. 2017;525(9):2202–2215. doi: 10.1002/cne.24199. https://doi.org/10.1002/cne.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel E-M, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Human Molecular Genetics. 2001;10(4):329–338. doi: 10.1093/hmg/10.4.329. https://doi.org/10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(5):2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Fatty acylation of proteins: The long and the short of it. Progress in Lipid Research. 2016;63:120–131. doi: 10.1016/j.plipres.2016.05.002. https://doi.org/10.1016/j.plipres.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JE, Mankowski JB, Sideris J, Goldman BD, Hatton DD, Mirrett PL, … Bailey DB. Trajectories and predictors of the development of very young boys with fragile X syndrome. Journal of Pediatric Psychology. 2009;34(8):827–836. doi: 10.1093/jpepsy/jsn129. https://doi.org/10.1093/jpepsy/jsn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, … Holt CE. Dynamic axonal translation in developing and mature visual circuits. Cell. 2016;166(1):181–192. doi: 10.1016/j.cell.2016.05.029. https://doi.org/10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackpole EE, Akins MR, Fallon JR. N-myristoylation regulates the axonal distribution of the Fragile X-related protein FXR2P. Molecular and Cellular Neuroscience. 2014;62:42–50. doi: 10.1016/j.mcn.2014.08.003. https://doi.org/10.1016/j.mcn.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harbor Perspectives in Biology. 2013;5(1):a007898. doi: 10.1101/cshperspect.a007898. https://doi.org/10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepniak B, Kästner A, Poggi G, Mitjans M, Begemann M, Hartmann A, … Ehrenreich H. Accumulated common variants in the broader fragile X gene family modulate autistic phenotypes. EMBO Molecular Medicine. 2015;7(12):1565–1579. doi: 10.15252/emmm.201505696. https://doi.org/10.15252/emmm.201505696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanini F, Unen LV, Bakker C, Sacchi N, Galjaard H, Oostra BA, Hoogeveen AT. Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P. Biochemical Journal. 1999;343(Pt 3) [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H. Protein myristoylation in protein–lipid and protein–protein interactions. Biophysical Chemistry. 1999;82(2–3):129–137. doi: 10.1016/s0301-4622(99)00112-x. https://doi.org/10.1016/S0301-4622(99)00112-X. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. The Journal of Neuroscience. 2009;29(15):4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. https://doi.org/10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Wu J, Tai HC, Schuman EM. Axonal translation of β-catenin regulates synaptic vesicle dynamics. The Journal of Neuroscience. 2013;33(13):5584–5589. doi: 10.1523/JNEUROSCI.2944-12.2013. https://doi.org/10.1523/JNEURO-SCI.2944-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, … Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. The Journal of Neuroscience. 2005;25(4):778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. https://doi.org/10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, … Darnell RB. A mouse model of the human fragile X syndrome I304N mutation. PLoS Genetics. 2009;5(12):e1000758. doi: 10.1371/journal.pgen.1000758. https://doi.org/10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, O’Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. The EMBO Journal. 1995;14(21):5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer SE, Doll SG, Garcia ADR, Akins MR. Splice form-dependent regulation of axonal arbor complexity by FMRP. Developmental Neurobiology. 2016 doi: 10.1002/dneu.22453. https://doi.org/10.1002/dneu.22453. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.