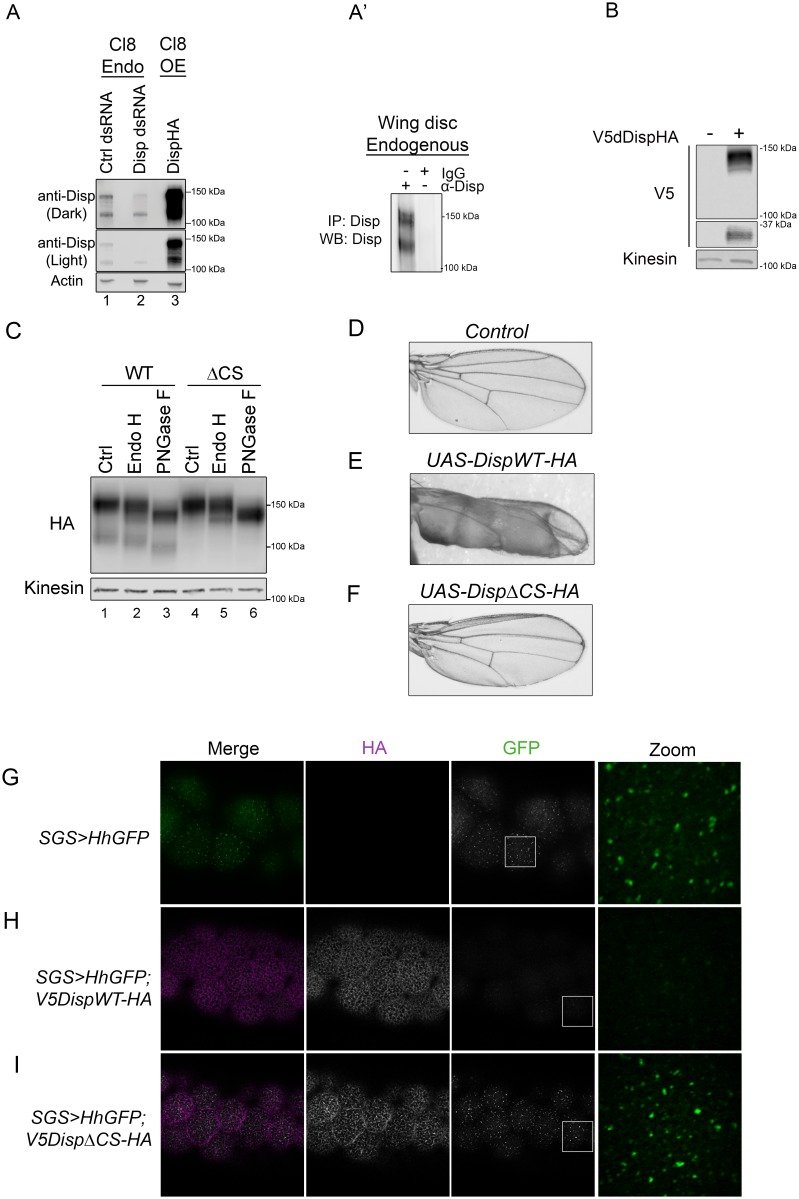
Figure 5. Cleavage is required for Disp activity in vivo.
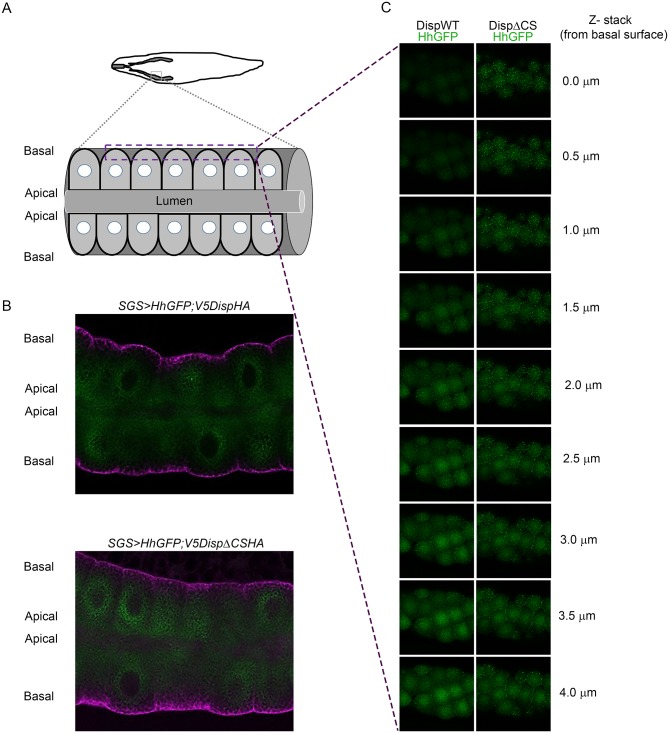
(A) Lysates from Drosophila Cl8 cells treated with control or disp dsRNA or transfected with pAc-dispHA were analyzed by western blot using anti-dDisp. Actin is the loading control. (A’) Endogenous dDisp150 and dDisp110 were specifically immunoprecipitated with anti-dDisp from wing imaginal disc lysate. (B) V5dDispHA was expressed in Cl8 cells and lysates were analyzed by western blot to confirm generation of the V5 fragment. Kinesin is the loading control. (C) Lysates were prepared from Cl8 cells expressing Δ206–238 (ΔCS) dDispHA protein and analyzed by western blot. Lysates were treated with Endo H or PNGase F. Kinesin is the loading control. (D–F) Wild type or ΔCS dDispHA proteins were expressed dorsally in wing imaginal discs using apterous-GAL4. Representative male wings are shown. (G–I) WT or ΔCS V5dDispHA proteins (magenta) were expressed with HhGFP (green) in salivary glands using SGS-GAL4. Maximum intensity projections of basolateral and basal optical sections of salivary glands are shown. Square in the GFP images indicates zoom area.