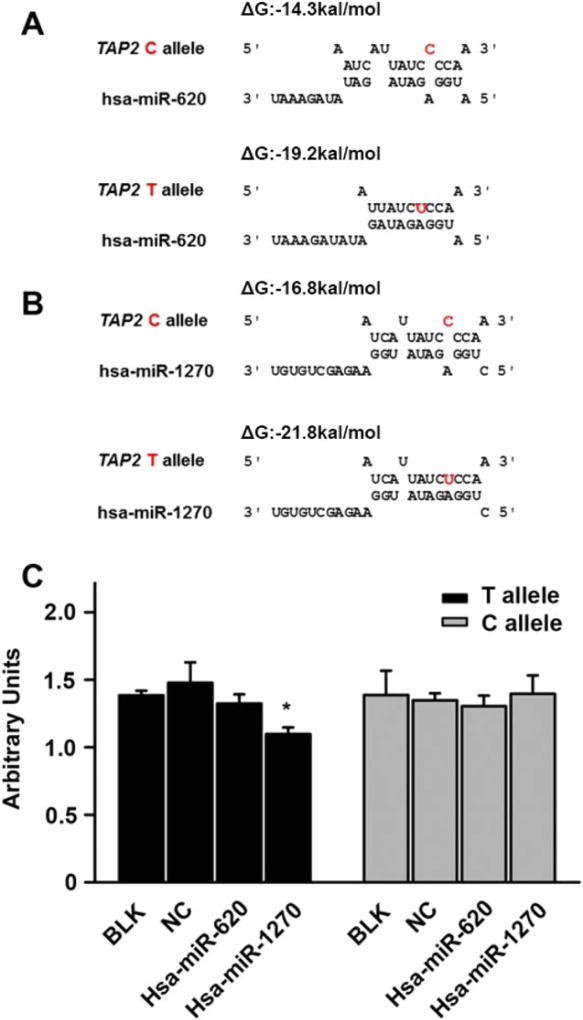
Fig. 3.
The hsa-miR-1270 suppressed reporter gene expression in an allele special manner. (A) Targeting prediction of hsa-miR- 620 to the 3′-UTR of TAP2 based on rs241456 allele. The free energy for binding of the miRNA to the T allele is stronger than the C allele. (B) Targeting prediction of hsa-miR-1270 to the 3′ -UTR of TAP2 based on rs241456 allele. The free energy for binding of the miRNA to the T allele is stronger than the C allele. (C) In HEK 293 cells, using luciferase reporter assay to detect the effect of each allele of rs241456. After HEK 293 cells were transiently transfected with a plasmid containing either the T or C allele for rs241456 together with hsa-miR-1270 mimics, hsa-miR-620 mimics, or an miRNA-negative control (NC). Renilla luciferase was measured as an internal control for the transfection efficiency, and firefly luciferase was measured to compare the gene expression levels. All experiments were carried out in triplicate. *P <0.05.