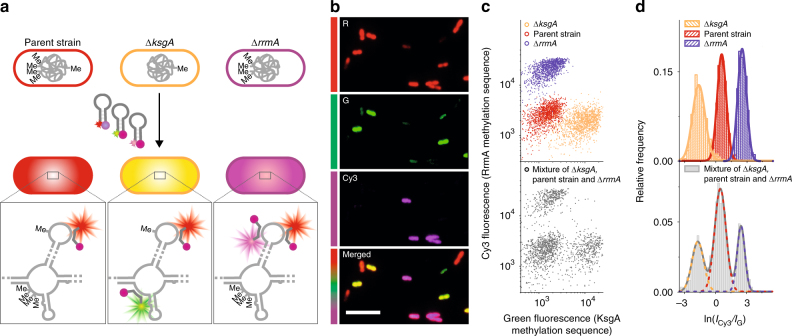
Fig. 4.
Multiplexed detection of rRNA methylations with MR-FISH. a Multiplexed assay for two rRNA methylations, using three Molecular Beacons, staining an unmethylated portion of the 23S rRNA (Alexa Fluor 647-labeled), the KsgA methylation sequence (Alexa Fluor 488-labeled) and the RrmA methylation sequence (Cy3-labeled). Three strains of E. coli are used: one expressing active KsgA and RrmA (parent strain), one mutant strain with the KsgA methyltransferase deleted from its genome (ΔksgA) and another mutant strain with the RrmA methyltransferase deleted from its genome (ΔrrmA). b False-colored fluorescence images of a mixture of parent strain, ΔksgA and ΔrrmA E. coli stained with the cocktail of Molecular Beacons. Scale bar: 10 μm. c Scatter plots of green and Cy3 fluorescence intensities of parent strain (n = 1362 cells), ΔksgA (n = 1411 cells) and ΔrrmA (n = 1288 cells) bacteria assayed separately and in a mixture (n = 1659 cells). d Log-normally distributed ratios of Cy3 fluorescence intensity (ICy3) to green fluorescence intensity (IG) of bacteria stained by MR-FISH in (c)