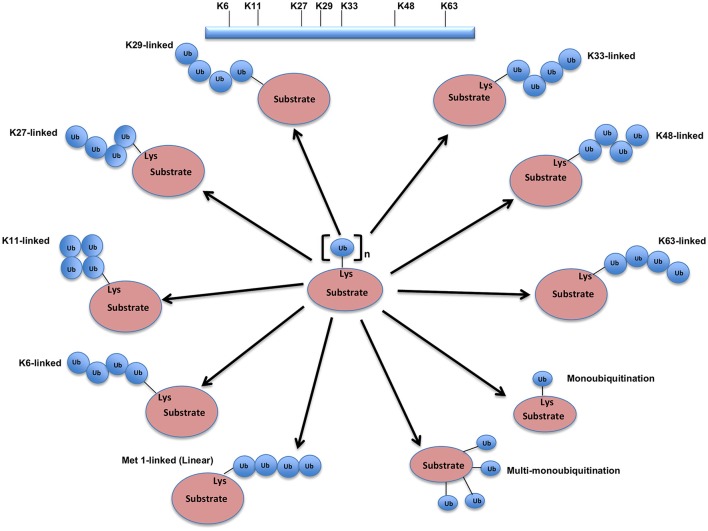
Figure 2.
Different types of Ubiquitination and specific function of Ubiquitin linkages. Ubiquitination process can be broken down into three groups, based on how the substrate protein gets tagged by ubiquitin molecule(s). Modification of the substrate with the formation of an isopeptide bond between the C-terminus of the activated ubiquitin and a single substrate lysine leads to the monoubiquitination, when multiple substrates' lysine residues modify with a single ubiquitin molecule each, that results into the multi-monoubiquitination and polyubiquitin chains using one of the seven available lysine residues (K6, K11, K27, K29, K33, K48, or K63) of one ubiquitin molecule attached via an isopeptide bond to the C-terminus of another ubiquitin molecule add further complexity to ubiquitin-encoded protein fate. K48-linked polyubiquitinated proteins are known to be highly unstable and call for degradation via 20S proteasome both for normally folded or misfolded proteins otherwise. K29- and K33-branched mixed chains have shown to regulate AMP-activated protein kinase-related kinases. Polyubiquitination in vivo through K29/K33-linked mixed chains blocks their kinase activation by interfering with phosphorylation of the activation-loop residues. K29-branched ubiquitin chains also shown to promote the proteasomal and lysosomal degradation of proteins, whereas K63-branched polyubiquitination majorly plays a key role in various non-degradative processes such as regulation of endocytosis, DNA repair, protein kinase activation, signal transduction, intracellular trafficking of membrane proteins, and stress responses. K63 mediated linkages also known to facilitate the autophagic degradation of substrate proteins and their associated cellular materials, such as damaged mitochondria and invading pathogens. Monoubiquitination and multi-monoubiquitination have been implicated in non-proteasomal regulatory functions like proteins translocation to the nucleus, cytoskeleton and endocytic machinery, virus budding, DNA repair, or modulating enzymatic activity and protein-protein interactions. Majority of the ubiquitin attachment on the protein appears to be at lysine residue, although N-terminal methionine (M1) and cysteine modifications have also been reported. Formation of a peptide bond between the N-terminal methionine residue of one ubiquitin molecule and the C-terminal glycine of the next in the chain results into the linear ubiquitin chains. Linear ubiquitin chains i.e. M1-linked chains primarily play pivotal roles in inflammatory and immune responses. Linear ubiquitin chain is formed by LUBAC (linear ubiquitin chain assembly complex), a multisubunit member of RBR family of E3 ligases. The complex is made of three enzymes: HOIP, HOIL1, and SHARPIN.