Abstract
MicroRNAs (miRNAs) are among the class of noncoding small RNA molecules and play a crucial role in post-transcriptional regulation in plants. Although Lilium is one of the most popular ornamental flowers worldwide, however, there is no report on miRNAs identification. In the present study, therefore, miRNAs and their targets were identified from flower, leaf, bulblet and bulb of Lilium lancifolium Thunb. by high-throughput sequencing and bioinformatics analysis. In this study, a total of 38 conserved miRNAs belonging to 17 miRNA families and 44 novel miRNAs were identified. In total, 366 target genes for conserved miRNAs and 415 target genes for novel miRNAs were predicted. The majority of the target genes for conserved miRNAs were transcriptional factors and novel miRNAs targeted mainly protein coding genes. A total of 53 cleavage sites belonging to 6 conserved miRNAs families and 14 novel miRNAs were identified using degradome sequencing. Twenty-three miRNAs were randomly selected, then, their credibility was confirmed using northern blot or stem-loop qRT-PCR. The results from qRT-PCR analysis showed the expression pattern of 4 LL-miRNAs was opposite to their targets. Therefore, our finding provides an important basis to understand the biological functions of miRNAs in Lilium.
Introduction
MicroRNAs (miRNAs) are a class of 20–24 nucleotide (nt) noncoding small RNA molecules and play a crucial role in post-transcriptional regulation in animals and plants1,2. In plants, microRNA genes are transcribed by RNA polymerase II into primary miRNAs (pri-miRNAs) with a cap and a poly(A) tail. The pri-miRNAs are then processed into hairpin precursors (pre-miRNAs) by a protein complex consisting of the Dicer-like 1 (DCL1), the C2H2-zinc finger protein SERRATE 11(SE), and the double-stranded RNA-binding protein HYPONASTICLEAVES1 (HYL1)3. The miRNA duplexes (miRNA/miRNA*) are released from pre-miRNAs by DCL1 and each strand in the miRNA duplex is methylated3. The miRNA strand is loaded into the ARGONAUTE (AGO) protein of RNA-induced silencing complex (RISC) to carry out its function1,3. Several research evidences revealed that miRNAs play important roles in diverse biological processes including plant growth, development, biotic and abiotic stress responses, and signal transduction4–6.
The first miRNA, lin-4, was identified from Caenorhabditis elegans in 19937. In plant, the first miRNAs were identified from Arabidopsis8. Following that, some miRNAs have been identified from plants using cloning or bioinformatics prediction9–12. The high-throughput sequencing technology was firstly used to identify A.thaliana miRNAs in 200513. Since then, thousands of miRNAs from different species have been discovered by high-throughput sequencing technology14–19. To date, a total of 35,828 mature miRNAs sequences from 223 different species (ranging from viruses to humans) have been identified according to the miRBase database (release 21, June 2014). However, there are only few researches conducted on miRNAs identification in ornamental flowers, including Phalaenopsis aphrodite, Rosa hybrida, Aquilegia coerulea and lotus japonicas20–23.
Lilium is a genus of Liliaceous perennial bulb plants. Lily (Lilium spp.), owing to their large and colorful flowers, have become one of the most popular ornamental flowers worldwide24. In addition to their ornamental value, some lily species are edible and has long been used as traditional medicine in China and Korea25–28. The genus Lilium has more than 100 species worldwide, of which 55 species and 18 varieties are originated in China29. Lilium lancifolium Thunb. is a well-known lily species widely distributed in China, and is often planted for landscaping design. It is one of Lilium species formed Asiatic hybrid lilies by interspecific crosses and give them the characteristics of orange flower and raised spot in petal30. Several studies have revealed its better capacity for resisting high and low temperature, drought, disease and changing soil salinity than other lilies31,32. Therefore, it has been used to produce progeny with desirable stress resistance in lily hybrid breeding32. Although several research reports shown that miRNAs play crucial roles in plant growth, development and response to stress in plant, to the best of our knowledge there is currently no report on Lilium miRNAs. In this study, therefore, we employed the high-throughput sequencing and bioinformatics analysis to identify miRNAs and their targets in L. lancifolium.
Results
Construction and sequencing of small RNA libraries
To identify the miRNAs in Lilium, total RNAs were extracted from flower, leaf, bulblet and bulb of L. lancifolium and then used to construct four small RNA libraries (Supplementary Table S1). Then the four small RNA libraries were sequenced using Illumina HiSeq. 2500 sequencing platform and analyzed in bioinformatics. A total of 19,025,905 raw reads from flower, 19,636,648 from leaf, 22,776,684 from bulblet, and 18,676,061 from bulb were obtained. After removing adaptors, low quality reads and contaminants, 14,090,897clean reads from flower, 15,279,574 from leaf, 17,560,878 from bulblet and 14,935,394 from bulb were obtained (Table 1). The clean reads and unique reads of four tissues were subjected to analysis of the size distribution as shown in Fig. 1. The majority of the clean reads of small RNAs in four samples were 21 to 26 nt in size. The 21 nt class was the most abundant in flower and bulb, followed by 22, 24 and 26 nt classes (Fig. 1A). In leaf and bulblet 23 nt small RNAs are the most frequent, followed by 26, 22 and 24 nt. However, the 24 nt peak is found to be dominant at a unique read level in all four samples (Fig. 1B).
Table 1.
Statistics of sequencing reads from flower, leaf, bulblet and bulb libraries of L. lancifolium.
| Samples | Raw reads | Clean reads (18–30 nt) | Unique reads | Mapped reads | rRNA/tRNA/ snRNA/snoRNA | Without annotation |
|---|---|---|---|---|---|---|
| Flower | 19,025,905 | 14,090,897 | 1,710,130 | 9,552,373 | 8,453,061 | 5,634,868 |
| Leaf | 19,636,648 | 15,279,574 | 1,103,780 | 10,493,835 | 10,263,031 | 5,012,994 |
| Bulblet | 22,776,684 | 17,560,878 | 1,394,751 | 12,803,064 | 11,776,773 | 5,764,221 |
| Bulb | 18,676,061 | 14,935,394 | 1,965,587 | 88,931,80 | 7,156,402 | 7,774,593 |
Figure 1.
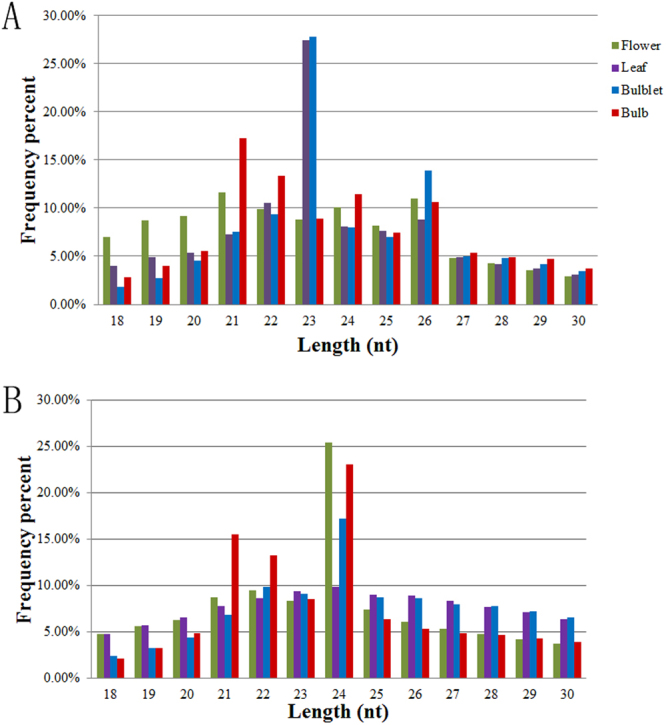
The length distribution of the clean and unique reads from flower, leaf, bulblet and bulb of L. lancifolium. (A) Clean reads; (B) Unique reads.
Conserved miRNAs in L. lancifolium
To analyze the population of conserved miRNAs in L. lancifolium, sRNA sequences of the four libraries were compared with known mature miRNAs from other plants and Lilium RNA sequence. A total of 38 conserved miRNAs precursors were identified from four sRNA datasets, as shown in Table 2 and Supplementary Table S2. All these conserved miRNAs belong to 17 miRNA families and in most miRNA families, more than one precursors were identified. Among them, MIR156 and MIR159 were the largest families identified with five members, followed by MIR166, in which four members were identified. However, several miRNA families possessed only one precursor, including MIR162, MIR167, MIR172, MIR390, MIR399, MIR408 and MIR845. In most miRNA families, at least one conserved miRNA precursor with miRNA* from small RNA sequencing was identified (Supplementary Fig. S1).
Table 2.
Conserved miRNAs identified from flower, leaf, bulblet and bulb libraries of L. lancifolium.
| Family | miRNA | Number of precursor sequence | Sequence (5′-3′) | Length (nt) | Clean reads | |||
|---|---|---|---|---|---|---|---|---|
| Bulblet | Bulb | Flower | Leaf | |||||
| MIR156 | LL-miR156a | c45184.graph_c0 | ugacagaagagagugagcac | 20 | 263 | 29 | 32 | 2 |
| c51027_g1 | ||||||||
| c103364_g2 | ||||||||
| c205656_g1 | ||||||||
| LL-miR156i | c45184.graph_c0 | ugacagaaagaguagugagca | 21 | 8 | 1 | 2 | 1 | |
| MIR159 | LL-miR159a | CL2574.Contig1_All | uggauugaagggagcucuaca | 21 | 11 | 13 | 2 | 4 |
| CL2574.Contig4_All | ||||||||
| LL-miR159b | c117583_g1 | uuuggauugaagggagcucua | 21 | 1446 | 969 | 671 | 379 | |
| c44060.graph_c0 | ||||||||
| LL-miR319a | c120927_g2 | uuggacugaagggagcucccu | 21 | 1 | 2 | 0 | 0 | |
| MIR160 | LL-miR160a | c19797.graph_c0 | ugccuggcucccuguaugcca | 21 | 3 | 1 | 8 | 3 |
| c50045_g1 | ||||||||
| LL-miR160f | c99102_g1 | cugccuggcucccugaaugcc | 21 | 1 | 18 | 2 | 3 | |
| MIR162 | LL-miR162a | Unigene26825_All | ucgauaaaccucugcauccgg | 21 | 17 | 52 | 50 | 4 |
| MIR164 | LL-miR164a | c96196_g1 | uggagaagcagggcacgugca | 21 | 11 | 0 | 18 | 0 |
| c30311.graph_c0 | ||||||||
| MIR166 | LL-miR166f | Unigene32510_All | ucucggaccaggcuucauucc | 21 | 49 | 40 | 366 | 6 |
| c39808.graph_c0 | ||||||||
| c121414_g2 | ||||||||
| LL-miR166g | c106540_g1 | ucggaccaggcuucauuccuc | 21 | 369 | 122 | 259 | 92 | |
| MIR167 | LL-miR167a | c105919_g1 | ugaagcugccagcaugaucuga | 21 | 7 | 37 | 1923 | 132 |
| MIR168 | LL-miR168a | c71740.graph_c0 | ucgcuuggugcaggucgggaa | 21 | 40 | 137 | 6 | 4 |
| c24174.graph_c0 | ||||||||
| c105404_g1 | ||||||||
| MIR172 | LL-miR172a | c79803.graph_c0 | agaaucuugaugaugcugcaa | 21 | 0 | 0 | 24 | 0 |
| MIR390 | LL-miR390a | c7928.graph_c0 | aagcucaggagggauagcgcc | 21 | 4 | 0 | 16 | 0 |
| MIR395 | LL-miR395a | c12817.graph_c0 | ugaaguguuugggggaacucc | 21 | 208 | 1183 | 105 | 140 |
| LL-miR395k | c5100.graph_c0 | ugaagcguuugggggaacucc | 21 | 0 | 2 | 1 | 1 | |
| MIR396 | LL-miR396a | c103053_g1 | uuccacagcuuucuugaacug | 21 | 33 | 123 | 19 | 15 |
| Unigene25901_All | ||||||||
| LL-miR396f | c30849_g1 | uuccacggcuuucuugaacua | 21 | 38 | 78 | 81 | 5 | |
| MIR398 | LL-miR398b | c221661_g1 | uguguucucaggucaccccug | 21 | 55 | 70 | 11 | 71 |
| c35683_g1 | ||||||||
| MIR399 | LL-miR399a | c191701_g1 | ugccaaaggagacuugcccug | 21 | 2 | 0 | 3 | 5 |
| MIR408 | LL-miR408b | c121837_g2 | ugcacugccucuucccuggcu | 21 | 7 | 0 | 2 | 14 |
| MIR845 | LL-miR845 | c54746.graph_c0 | cgcucugauaccacuuguugg | 21 | 1 | 8 | 1 | 0 |
| MIR2118 | LL-miR2118e | c28582_g1 | uucccaaugccucucaugccaa | 22 | 2 | 0 | 10 | 1 |
| LL-miR2118a | Unigene6944_All | uugccgauaccacccauaccga | 22 | 4 | 6 | 3 | 0 | |
The sRNA sequencing results indicated that the clean reads of conserved miRNAs ranged from 1 up to more than 1,000 in 4 samples. Of all conserved miRNAs, the clean reads of MIR159, MIR167 and MIR395 exceeded 1,000 in one tissue. The clean reads of four miRNA families (MIR156, MIR166, MIR168 and MIR396) ranged from 100 to 1,000 at least in one tissue. However, the other miRNA families (MIR160, MIR162, MIR164, MIR172, MIR390, MIR398, MIR399, MIR408, MIR845 and MIR2118) had fewer than 100 reads in all four tissues (Table 2).
In a broader evolutionary context, L. lancifolium miRNAs were aligned to those of 13 other plants, including 11 monocotyledons (Aegilops tauschii, Brachypodium distachyon, Elaeis guineensis, Festuca arundinacea, Hordeum vulgare, Oryza sativa, Sorghum bicolor, Saccharum officinarum, Saccharum sp., Triticum aestivum and Zea mays) and 2 dicotyledons (Arabidopsis thaliana and Glycine max). Of the 17 L. lancifolium miRNA families, 15 were conserved in more than 6 plant species. These miRNAs were considered as well-conserved miRNA families. However, LL-miR845 and LL-miR2118 from L. lancifolium were found in only two and one plant species, respectively (Supplementary Table S2).
Novel miRNAs in L. lancifolium
To identify novel miRNAs that may be specific to L. lancifolium, all unannotated sRNAs were searched against the unigenes from Lilium transcriptome sequencing and EST from NCBI database using miRDeep2. After searching for potential precursors (pre-miRNAs) and predicting their stem-loop hairpin secondary structures, a total of 44 novel miRNAs were identified in four libraries. The novel miRNA sequences ranged in length from 20–24 nt. However, the sequences of most novel miRNAs were 21 nt length and started with a 5′-U (Table 3). The pre-miRNAs ranged in length from 127–1475 nt. The average minimum folding free energy value of the hairpin structures was −152 kcal/mol in L. lancifolium (Supplementary Table S3), which is higher than −76.8 kcal/mol found in Arabidopsis33. The structures of 44 novel miRNA precursors are shown in Supplementary Fig. S2. Nineteen miRNA* sequences from small RNA sequencing were discovered in these novel miRNA precursors. Half of novel miRNAs had more than 10 clean reads in at least one tissue. Only LL-miR09, LL-miR23 and LL-miR25 had more than 100 clean reads in leaf, bulb or flower tissue (Table 3). It is very interesting that two highly similar novel miRNAs, LL-miR07 and LL-miR14, were identified. Only two different bases were found between LL-miR07 and LL-miR14 mature miRNA sequences (Table 3). Moreover, the similarity of precursors between LL-miR07 and LL-miR14 was more than 85%.
Table 3.
Novel miRNAs identified from flower, leaf, bulblet and bulb libraries of L. lancifolium.
| miRNA | Number of precursor sequence | Sequence (5′-3′) | Length (nt) | Clean reads | |||
|---|---|---|---|---|---|---|---|
| Bulblet | Bulb | Flower | Leaf | ||||
| LL-miR01 | c33038.graph_c0 | uaguaaguuugcagagcagag | 21 | 0 | 0 | 53 | 0 |
| LL-miR02 | c1069.graph_c1 | cuugugcuucuggacugcucc | 21 | 0 | 56 | 0 | 0 |
| LL-miR03 | c15590.graph_c0 | aagguauagagucagacacuu | 20 | 0 | 0 | 9 | 0 |
| LL-miR04 | c25608.graph_c0 | agacgaucgcaccaaacuggcuau | 24 | 13 | 67 | 1 | 0 |
| LL-miR05 | c28639.graph_c0 | uuuucuaugucacucaauccaa | 22 | 0 | 2 | 3 | 0 |
| LL-miR06 | c30876.graph_c0 | aguaaguugagaagaguaggagaa | 24 | 3 | 3 | 0 | 1 |
| LL-miR07 | c31471.graph_c0 | uucacugccaccauccgccugu | 22 | 1 | 9 | 28 | 1 |
| LL-miR08 | c35378.graph_c0 | cgguugcuuagcuuguacucu | 21 | 0 | 12 | 1 | 0 |
| LL-miR09 | c36638.graph_c0 | ugcaccuccuccuccuuuucu | 21 | 56 | 13 | 33 | 244 |
| LL-miR10 | CL3742.Contig2_All | gguuugaugaaucugagcauc | 21 | 14 | 19 | 55 | 18 |
| LL-miR11 | c53683.graph_c0 | uuacgugucccuuaaucugacggg | 24 | 0 | 3 | 0 | 1 |
| LL-miR12 | c56352_g1 | gcucggguuaacggggaagug | 21 | 9 | 56 | 0 | 0 |
| LL-miR13 | c59256_g1 | uaugaaguuauauagguuguccgg | 24 | 1 | 6 | 0 | 0 |
| LL-miR14 | c79455.graph_c0 | uuuacugccaccauccgccugc | 22 | 3 | 12 | 5 | 0 |
| LL-miR15 | c94000_g1 | aagguaagagaaucaacaagaggu | 24 | 0 | 8 | 0 | 0 |
| LL-miR16 | c96112_g1 | uaggcaacaaauuagagucucu | 22 | 8 | 8 | 33 | 50 |
| LL-miR17 | c99691_g1 | uuuguauggucuguugaaauu | 21 | 4 | 0 | 0 | 0 |
| LL-miR18 | c109150_g1 | caggcggcgaggauggggaug | 21 | 2 | 5 | 0 | 0 |
| LL-miR19 | c109175_g1 | uugcuuagcuuguacucucgc | 21 | 1 | 4 | 4 | 0 |
| LL-miR20 | c110752_g2 | ugaaaauguagcacuagcacc | 21 | 1 | 7 | 0 | 0 |
| LL-miR21 | c111855_g1 | uugagaguagagagccaggug | 21 | 0 | 12 | 1 | 4 |
| LL-miR22 | c114504_g1 | aaaugaugaaucugagccuc | 20 | 8 | 0 | 10 | 6 |
| LL-miR23 | c114692_g4 | uagaggcgaugaugaugaaau | 21 | 44 | 795 | 75 | 119 |
| LL-miR24 | c117497_g1 | ugaagacuuggcaaccgacauc | 22 | 4 | 20 | 2 | 0 |
| LL-miR25 | c117720_g1 | ucugcccugauaugagcuccag | 22 | 36 | 0 | 137 | 0 |
| LL-miR26 | c117786_g3 | ucugaauagcaaacccaauuc | 21 | 3 | 5 | 1 | 0 |
| LL-miR27 | c166092_g1 | aaacgaucgauaaaccucugc | 21 | 0 | 4 | 2 | 0 |
| LL-miR28 | c48903.graph_c0 | aaugagaagacuagugacaagauu | 24 | 4 | 73 | 0 | 0 |
| LL-miR29 | CL711.Contig2_All | uucccuucggcugcaaauagc | 21 | 77 | 25 | 47 | 33 |
| LL-miR30 | CL719.Contig1_All | uagaggcgaugaugaugaaau | 21 | 0 | 2 | 0 | 1 |
| LL-miR31 | CL1297.Contig2_All | aucuuuggccuggagauagagg | 22 | 0 | 3 | 0 | 0 |
| LL-miR32 | c71927_g1 | ugugccaugcugugugcgucc | 21 | 2 | 30 | 2 | 0 |
| LL-miR33 | CL4047.Contig1_All | ugccgggcuaagauacaaggau | 22 | 1 | 0 | 2 | 1 |
| LL-miR34 | HM045458.1 | ucuauaugacucucggcaacgg | 22 | 0 | 1 | 25 | 2 |
| LL-miR35 | JZ391002 | uccaaagucagugaggggagc | 21 | 0 | 9 | 0 | 0 |
| LL-miR36 | Unigene13110_All | uucgagugacauauggaaacu | 21 | 1 | 3 | 0 | 0 |
| LL-miR37 | Unigene18554_All | ucaaucuuuggccuggagauagag | 24 | 2 | 10 | 4 | 0 |
| LL-miR38 | c68386.graph_c0 | ugggucuccucucauuccaug | 21 | 9 | 13 | 0 | 0 |
| LL-miR39 | Unigene25443_All | uucgagugacauauggaaacu | 21 | 1 | 3 | 0 | 0 |
| LL-miR40 | c51021.graph_c0 | ucaaagacgaaucugagcaua | 21 | 2 | 6 | 0 | 0 |
| LL-miR41 | c56504.graph_c0 | ucguaucugugguuugcuccu | 21 | 0 | 1 | 2 | 0 |
| LL-miR42 | c59249.graph_c0 | ugcaguuugguuuguggugug | 21 | 1 | 3 | 0 | 1 |
| LL-miR43 | GW589960 | cugucgagcuuccauacuggc | 21 | 0 | 3 | 0 | 0 |
| LL-miR44 | JZ391211 | uggaucuugaaccaaguguuc | 21 | 0 | 2 | 10 | 0 |
Prediction of miRNA targets
Plant miRNAs play important roles in diverse biological processes by cleaving target mRNAs or suppressing the translation of target genes. In order to understand the biological functions of L. lancifolium miRNAs, TargetFinder and psRNA Target software was used to predict putative target genes of novel and conserved miRNAs. The results from analysis showed that 366 target genes for 17 conserved miRNA families and 415 target genes for 40 novel miRNAs were predicted (Supplementary Tables S4 and S5). The majority of the target genes for conserved miRNAs were transcriptional factors, and many target genes were conserved between Lilium and other plant, such as squamosa promoter-binding proteins (SPL), MYB, proliferating cell factors (PCF), auxin response factor (ARF), DCL1, NAC domain transcription factor, cup-shaped cotyledon 2(CUC2), PHB, AGO1, APETALA, ATP sulfurylases, growth-regulating factors (GRF), UBC24 and blue copper protein, which were involved in various aspects of plant growth and development. However, some predicted target genes of several conserved miRNAs in L. lancifolium were different from those in other plants, including fasciclin-like arabinogalactan protein (FLA) and homeobox-leucine zipper protein HOX (MIR166), ethylene-responsive transcription factor RAP2-7 (MIR172), LRR receptor-like serine/threonine-protein kinase (MIR390), stellacyanin and cucumber peeling cupredoxin (MIR398). Interestingly many target genes for miR845 were predicted in L. lancifolium like patatin protein, phosphatidate cytidylyltransferase, dnaJ protein, and LRR receptor-like serine/threonine-protein kinase (Supplementary Table S4). The target genes of novel miRNAs were mainly predicted to be protein coding genes, such as polyphenol oxidase, polycomb group protein FIE2, serine/threonine-protein kinase, pentatricopeptide repeat-containing protein, DRM-type DNA-methyltransferase, ATP-dependent DNA helicase, F-box protein and so on (Supplementary Table S5).
Validation of miRNAs using northen hybridation and stem-loop RT-PCR
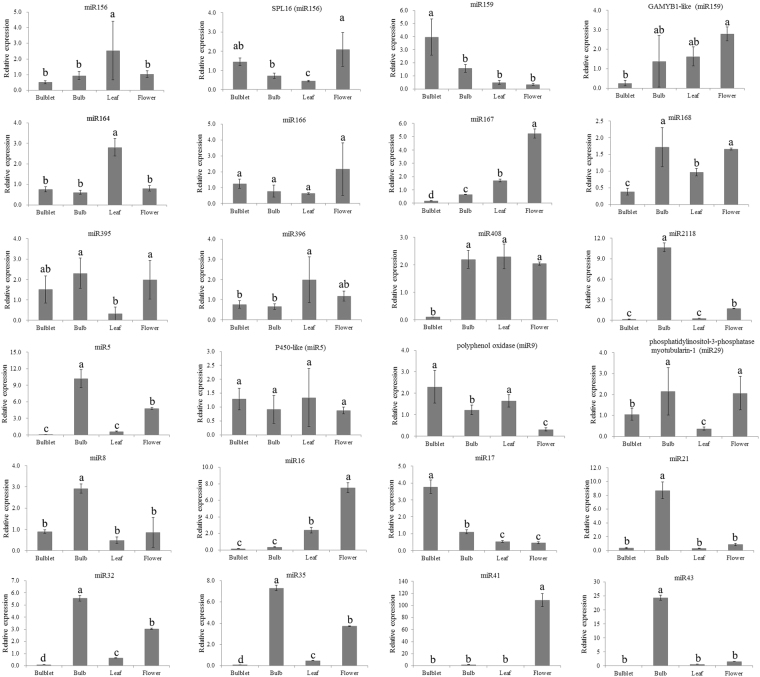
To confirm the credibility of miRNAs from the high-throughput sequencing and bioinformatics analysis, 4 novel miRNAs (LL-miR7, LL-miR9, LL-miR14 and LL-miR29) were randomly selected and their expressions in flower, leaf, bulblet and bulb of L. lancifolium were detected using northern blot analysis. All of them were detected in at least one tissue (Fig. 2). Two lily cultivars, ‘Brunello’ and ‘White heaven’, were also used for northen hybridization. All 4 novel miRNAs were detected in ‘Brunello’ or ‘White heaven’. In addition, 19 miRNAs (10 conserved and 9 novel miRNAs) were randomly selected for stem-loop RT-PCR analysis. All of these miRNAs were detected from flower, leaf, bulblet and bulb of L. lancifolium. Among them, 7 miRNAs (LL-miR2118, LL-miR5, LL-miR8, LL-miR21, LL-miR32, LL-miR35 and LL-miR43) were found primarily in bulb. Three miRNAs (LL-miR167, LL-miR16 and LL-miR41) were expressed predominantly in flower (Fig. 3). Three miRNAs (LL-miR159, LL-miR156 and LL-miR164) were highly expressed in bulblet or leaf. For the specificity and sensitivity of northern hybridization and stem-loop qRT-PCR34,35, the identification of conserved and novel miRNAs in L. lancifolium was effective and credible.
Figure 2.
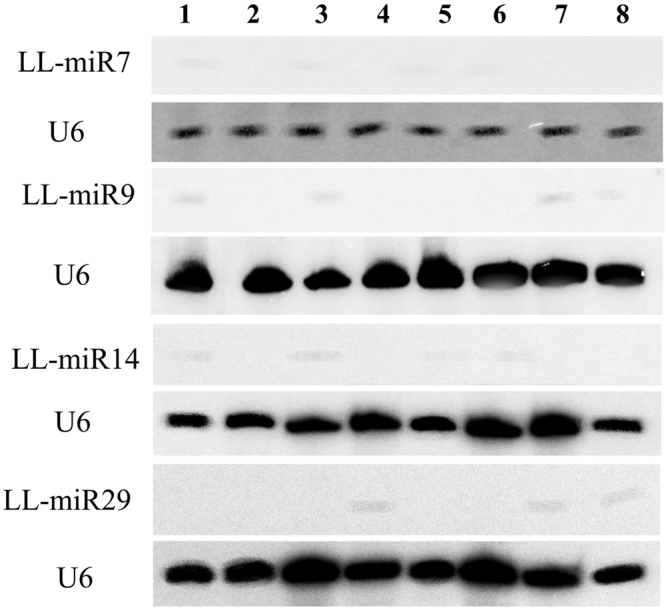
Detection of four novel miRNAs using Northern blotting. Lane 1–4, L. lancifolium; Lane 5,6, Brunello; Lane 7,8, White heaven; Lane1,5,7, flower; Lane 4,6,8, leaf; Lane2, bulblet; Lane3 bulb. U6 RNA is used as a loading control. The full-length blots are presented in Supplementary Fig. S3.
Figure 3.
Validation of miRNAs and targets using qRT-PCR. The X axis represents different tissues. The Y axis represents the relative expression level of miRNAs or targets.The amount of expression of miRNAs and targets was normalized to the level of 5.8S rRNA and 18S rRNA, respectively. Different letters indicate significant differences at P < 0.05 according to Duncan’s multiple range tests.
Verification of miRNA targets by degradome sequencing and RT-PCR
In order to validate the targets of conserved and novel miRNAs in L. lancifolium, the high-throughput degradome sequencing was used to detect the cleavage site of predicted targets. A total of 683,113,369 raw data were obtained. After removing adaptors, low quality reads and repeat sequences, 14,534,088 clean tags were yielded. A total of 12,194,038 clean tags were mapped perfectly to Lilium RNAs. After bioinformatics analysis, 27 and 26 cleavage sites were identified to 6 conserved miRNAs families and 14 novel miRNAs, respectively (Supplementary Tables S4 and S5). According to Addo-Quaye’s analysis method36, all targets identified by degradome sequencing could be classified into five categories: twenty-two cleavage sites with category 0, ten cleavage sites with category 1, nine cleavage sites with category 2, five cleavage sites with category 3, seven cleavage sites with category 4. Most cleavage sites (11/12) with category 3 and 4 were from novel miRNA targets. These targets of conserved miRNAs included SPLs, MYB, GRFs, DCL1, pentatricopeptide repeat-containing protein, Ras-group-related LRR protein, methyltransferase PMT7, chorismatemutase 3 and trifunctional UDP-glucose 4,6-dehydratase (Supplementary Table S4). Novel miRNAs could cleave P450-like, polyphenol oxidase, auxin response factor 7-like, Kunitz-type trypsin inhibitor, F-box protein, ferredoxin, FLS2, N-alpha-acetyltransferase 16, and so on (Supplementary Table S5).
To validate the regulation of target expression by miRNA, five targets for two conserved and three novel miRNAs were investigated using qRT-PCR. Among them, LL-miR9 could be detected in flower and bulb using northern blot. Polyphenol oxidase, a predicted target of LL-miR9, has higher expression in bublet and leaf. Phosphatidylinositol-3-phosphatase myotubularin-1 to potentially be targeted by LL-miR29 has decreased expression in leaf (Fig. 3). The result agreed with the expression of LL-miR29 from northern blot. In addition, LL-miR156 and LL-miR159 also had the opposite expression pattern with their targets (SPL16 and GAMYB1-like).
Discussion
Lilium species belong to genus Liliaceous, known as perennial herbaceous flowering plants growing from bulbs and among the most important cut flowers worldwide. Although several research reports shown that miRNAs play crucial roles in plant growth, development, and response to stress in plants, however, there is no report on Lilium miRNAs identification. The high-throughput sequencing is an effective method to screen miRNAs from various plants. Up to date, many miRNAs from ornamental plant, including Lycoris aurea, Nelumbo nucifera, Herbaceous peony, Cymbidium ensifolium, Ginkgo biloba, Prunus mume and rose21,37–42, have been identified using this method. To discover miRNAs in Lilium and understand their functions, we constructed and sequenced four small RNA libraries from flower, leaf, bulblet and bulb of L. lancifolium. The result of this research adds our knowledge to understand about the role of miRNAs from Liliaceae, in which a number of genera are popular cultivated plants with ornamental value.
The study on miRNAs in Lilium was limited to their large genome (~36 Gb) compared with other plants in which genome has been sequenced, such as Arabidopsis43, rice44, wheat45, Populus46. In this study, RNA sequences from Lilium in NCBI database, transcriptome data from Lilium pumilum and an Asiatic hybrid cultivar ‘Easy Dance’ besides to L. lancifolium were also used to predict miRNAs and their targets. As a result a total of 17 conserved miRNA families and 44 novel miRNAs were identified compared with only 9 conserved miRNA families and 17 novel miRNAs using L. lancifolium RNA. Therefore, we believe that deep sequencing of different tissues at various developmental stages could be necessary to fully disclose the miRNA function in Lilium. And in comparison to other close physiologically plants, the number of miRNAs identified from L. lancifolium are less than rice (713), wheat (119), maize (321), Brachypodium distachyon (525) which have been well genomic sequenced, but more than those plants without genome sequencing data, like Elaeis guineensis (6), Festuca arundinacea (15), Saccharum officinarum (16) (miRbase database release 21).
The lengths of plant sRNAs usually ranged from 21 nt to 24 nt47. Among them, 24 nt sRNA was the most abundant, followed by 21 nt class13,48,49. However, our research result shows that 21 nt sRNAs were more abundant in flower and bulb whereas 23 nt sRNA in leaf and bulblet were more abundant than 24 nt sRNA in L. lancifolium. It has been reported that the 21 nt sRNA is the most abundant sRNA species in Populus balsamifer, Chinese Wild Vitis pseudoreticulata and Pinus cordata48,50,51. Our results from flower and bulb supported the speculation given by Han and colleagues that the major sRNA species were 21 nt in perennial plants51. The 23 nt sRNAs could arising from loci dominated by 24 nt siRNAs52 and MIR genes53, but their biological functions are less understood. Therefore, a large number of 23 nt sRNA may play a special role in leaf and bulblet of L. lancifolium.
After analyzing miRNAs identified in this study, we found that the 5′ ends of most miRNAs were U (84%for conserved miRNAs and 70.8% for novel miRNAs) in L. lancifolium. The result was consistent with previous reports that the U at 5′end of miRNA favored the combination with AGO154. It has been reported that miRNAs had differential accumulation patterns in many plant species, and possessed their own precise regulation processes through the tissue dependent miRNA biogenesis in different plant species11,55. In this study, the expression analysis of miRNAs from qRT-PCR and northern blot showed that many conserved and novel miRNAs were tissue biased in L. lancifolium. This result suggested that the miRNAs might play very important roles in development of different tissue.
In order to understand the functions of miRNAs in L. lancifolium, a large number of targets of conserved and novel miRNAs were predicted in this research. These conserved miRNAs LL-miR156, LL-miR159, LL-miR319, LL-miR160, LL-miR164, LL-miR166, LL-miR172 and LL-miR396 targeted SPLs, MYBs, PCFs, ARFs, NACs, HD-ZIP III, AP2s and GRFs, respectively. Our result is in agreement with previous research that the targets of conserved miRNAs in plant are mainly transcription factors56. These conserved miRNAs and their transcription factor targets in lily might have similar functions on plant growth and development as those in other plant species. However, the LL-miR398 has been predicted to target type I blue copper proteins rather than reported CSD, CoX5b-1 and CCS1, which were involved in responses to environmental stresses57–59. In this study, 27 targets of LL-miR156, LL-miR159, LL-miR162, LL-miR390, LL-miR396 and LL-miR2118 have been validated by degradome sequencing. The result from qRT-PCR analysis showed that LL-miR156 and LL-miR159 had the opposite expression pattern with their targets.
The novel miRNAs were mainly predicted to be protein coding genes. Among 415 predicted targets, 26 were validated by degradome sequencing. Although some cleavage sites for predicted targets, which were matched very well with novel miRNAs, were not detected using degradome sequencing, it was possible that novel miRNAs regulated them at the level of translation. The novel LL-miR09 and LL-miR35 were predicted to target polyphenol oxidase, and the cleavage site of LL-miR09 has been identified by degradome sequencing. In Populus and Salvia miltiorrhiza, miR1444 and Smi-miR12112 have been reported to regulate a subset of polyphenol oxidases, which have important roles in plant development and response to biotic and abiotic stresses60–62. LRR receptor-like serine/threonine-protein kinase FLS2 and ERECTA were predicted as targets of LL-miR21 and miR35, respectively, and the cleavage site of FLS2 has been detected. The FLS2 has been reported to perceive the bacterial elicitor flagellin in Arabidopsis63. The ERECTA was involved in the thermo tolerance, stomatal development, plant architecture in Arabidopsis64. The serine/threonine-protein kinase ACR4 targeted by LL-miR25 plays important roles in cell division and differentiation in Arabidopsis65. The polycomb group protein FIE2, which prevents fertilization-independent seed development in Arabidopsis66, is a potential LL-miR08 target. The pentatricopeptide repeat-containing protein, which was potentially targeted by LL-miR18, LL-miR43 and LL-miR44, could affect chloroplast development in Arabidopsis67. DRM-type DNA-methyltransferase involved in RNA-directed DNA methylation in Arabidopsis, was predicted as target of miR2068. ATP-dependent DNA helicase DDM1 participated in UV-B induced and oxidative DNA damage repair in Arabidopsis69, was potentially targeted by LL-miR36. F-box protein potentially targeted by LL-miR26, LL-miR38 and LL-miR44, has been reported to mediate bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis70. It is very interesting that LL-miR07 and LL-miR14 owed highly similar mature miRNA and precursor sequences. Therefore, we speculated that LL-miR07 and LL-miR14 might be derived from the same ancestor. Both of them were predicted to target ARF7-like, which regulates lateral root formation, differential growth of hypocotyls in Arabidopsis and fruit set in tomato71,72. Since these novel miRNAs were predicted to participate in plant growth, development, biotic and abiotic stress responses, and signal transduction and so on, further studies are recommended to understand the functions of novel miRNAs in Lilium species.
Methods
Plant material and RNA preparation
L. lancifolium was selected as the experiment material, and grown in the greenhouse of Beijing University of Agriculture, Beijing, China. Flowers, leaves, bulblets and bulbs of L. lancifolium were collected and immediately frozen in liquid nitrogen. The frozen samples were then stored at −80 °C for future analysis. The TRIzol® reagent (Invitrogen, USA) was used to extract total RNA from four samples according to the manufacturer’s protocol. Finally, the integrity of total RNA was confirmed using 1% agarose gel electrophoresis and Agilent 2100 Bio analyzer (Agilent Technologies, USA).
Small RNA library construction and sequencing
Small RNA libraries of flowers, leaves, bulblets and bulbs were constructed using previously described methods73. Briefly, small RNAs fragments of 10–30 nt were purified from a 15% denaturing polyacrylamide gel and then ligated with 5′ and 3′ adapters. After being reverse-transcribed by Superscript II reverse transcriptase (Invitrogen) and amplified by PCR, about 20 µg products from each sample were sequenced using Illumina HiSeq. 2500 sequencing platform (Illumina Inc.; San Diego, CA, USA) at the Biomarker Technologies (Beijing, China).
Prediction of conserved and novel miRNA
After removal of chip adaptor sequences, low quantity reads and contaminations, the clean 18–30 nt small RNAs were mapped to GenBank (http://www.ncbi.nlm.nih.gov/) and Rfam (version 10.1) database (http://rfam.sanger.ac.uk) with a cut-off value of 0.01, and rRNA, tRNA, snRNA, snoRNA were removed to produce filtered small RNAs. All available Lilium RNA sequences were collected to predict miRNAs and their targets, including unigenes from transcriptome sequencing of L. lancifolium (SRA632698), Lilium pumilum (SRA633315), Asiatic hybrid lily cultivars ‘Easy Dance’ (SRA538278), and 3902 ESTs (Lilium formosanum, Lilium longiflorum, Lilium regale, Lilium hybrid division VII, and Lilium davidii var. willmottiae) downloaded from NCBI. All filtered small RNA, which were aligned against miRbase database (release 21) (http://www.mirbase.org/) with no more than two mismatches, were aligned against Lilium RNA sequences. Their flanking sequences were fold with mfold soft74. The filtered small RNA in perfect stem-loop structure was considered as conserved miRNAs. The miRDeep2 with modified parameter was used to identify novel miRNAs and check the secondary structures of putative pre-miRNAs75. The minimum free energy index (MFEI) was calculated using the equation: MFEI = AMFE/(G + C)%. The adjusted MFE (AMFE) represented the MFE of 100 nucleotides. It was calculated using (MFE/length of RNA sequence) × 10076.
Target prediction of conserved and novel miRNA
Lilium RNA sequences above for miRNA prediction were also used for target prediction. TargetFinder and psRNA Target were applied to predict the putative targets of conserved and novel miRNA77,78.
Quantitative real-time PCR analysis of miRNAs and targets
The stem-loop RT-PCR was used to validate the miRNAs from deep sequencing and to analyze their expression patterns. Total RNA of flower, leaf, bulblet, and bulb were extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instruction. Then, total RNA was reverse-transcribed to cDNA using stem-loop RT primer by the PrimeScript RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. All primers for stem-loop RT-RCR were designed according to the Chen’s report35 and listed in Supplementary Table S6. The qRT-PCR reactions were performed using the SYBR Premix Ex Taq II solution (TaKaRa, Dalian, China) as the following condition: 95 °C for 5 minutes, then 40 cycles of denaturation at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. For the qRT-PCR analysis of the miRNA targets, total RNA was used for synthesizing reverse transcripts using PrimeScript RT reagent Kit (Takara, Dalian, China) according to the manufacturer’s instructions. Specific primer pairs for miRNA targets were designed to amplify cDNA (Supplementary Table S6). qPCR was performed using SYBR Premix Ex Taq II (TaKaRa, Dalian, China) under the following conditions: 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. The qRT-PCR reactions were performed on the BIO-RAD iQ5 (Applied Biosystems, Foster City, CA). Each sample was processed in triplicate, and the relative expression were calculated using 2−ΔΔCT 79. The 5.8S rRNA80 and 18S rRNA were used as references to normalize the expression level of miRNAs and their targets. The data were statistically analyzed using SAS Version 9.0 software (SAS Institute, Cary, NC, USA) using Duncan’s multiple range test at the P < 0.05 level of significance.
Degradome library construction and target identification
To investigate the potential targets of conserved and novel miRNAs, a degradome library was constructed using mixture of mRNA from flowers, leaves, bulblets and bulbs of L. lancifolium according to the parallel analysis of the RNA ends protocol81, and sequenced using Illumina HiSeqTM 2500 sequencing platform (Illumina Inc.; San Diego, CA, USA) at the Beijing Genomics Institute (BGI) (Shenzhen, China). A Public software package, CleaveLand3.0 was used for analyzing sequencing data. All the putative target genes were used as queries to align against Lilium RNA sequences. The true miRNA cleavage sites from background noise were identified using a target plot82.
Northern blot analysis
Total RNA was isolated from plant tissues using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. For Northern blots, 40 µg of total RNA was separated by electrophoresis on 17% polyacrylamide gel and electrically transferred to nylon N+ membrane. Blots were hybridized with [ϒ-32P] ATP-labeled oligonucleotide probe. Hybridization signal intensity was measured using a PhosphorImager (GE Healthcare). Sequences of the oligonucleotide probes are listed in Supplementary Table S6.
Data availability
The raw data (Accession Number: SRA633909) in the study can be obtained from SRA database.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant NO. 31640007), the High-level Scientific Research Cultivation Project of BUA (GJB2015004), the Project of Construction of Innovative Teams and Teacher Career Development for Universities and Colleges Under Beijing Municipality (IDHT20150503), the Beijing Municipal Education Commission (CEFF-PXM2017_014207_000043).
Author Contributions
X.H. designed this study, performed the lab experiments, analyzed the data and wrote the manuscript. A.G.S. wrote the manuscript. S.X. designed this study, analyzed the data and wrote the manuscript. W.W. designed this study and wrote the manuscript. All authors have reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21193-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenhe Wang, Email: wwhals@163.com.
Shufa Xu, Email: xushufa@caas.cn.
References
- 1.Voinnet O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;2008:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Chen X. Small RNAs and Their Roles in Plant Development. Annu. Rev. Cell Dev. Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers K, Chen X. Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Guo H-S. MicroRNA Directs mRNA Cleavage of the Transcription Factor NAC1 to Downregulate Auxin Signals for Arabidopsis Lateral Root Development. Plant Cell Online. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4encodes small RNAs with antisense comlementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Plant Signal. Behav. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 10.Lu S. Novel and Mechanical Stress-Responsive MicroRNAs in Populus trichocarpa That Are Absent from Arabidopsis. Plant Cell Online. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He XF, Fang YY, Feng L, Guo HS. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 2008;582:2445–2452. doi: 10.1016/j.febslet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Arazi T, et al. Cloning and characterization of micro‐RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 13.Lu C, et al. Elucidation of the Small RNA Component of the Transcriptome. Science (80-.). 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 14.Ruby JG, et al. Large-Scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Bar M, et al. MicroRNA Discovery and Profiling in Human Embryonic Stem Cells by Deep Sequencing of Small RNA Libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burnside J, et al. Deep Sequencing of Chicken microRNAs. BMC Genomics. 2008;9:185. doi: 10.1186/1471-2164-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei B, et al. Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon (L.) Beauv. Funct. Integr. Genomics. 2009;9:499–511. doi: 10.1007/s10142-009-0128-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Xu Y, Huan Q, Chong K. Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics. 2009;10:449. doi: 10.1186/1471-2164-10-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao C-Z, et al. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.) BMC Plant Biol. 2010;10:3. doi: 10.1186/1471-2229-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao YT, et al. Identification and characterization of the microRNA transcriptome of a moth orchid Phalaenopsis aphrodite. Plant Mol. Biol. 2014;84:529–548. doi: 10.1007/s11103-013-0150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei, H. et al. Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS One8 (2013). [DOI] [PMC free article] [PubMed]
- 22.Puzey JR, Kramer EM. Identification of conserved Aquilegia coerulea microRNAs and their targets. Gene. 2009;448:46–56. doi: 10.1016/j.gene.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 23.De Luis A, et al. Two MicroRNAs Linked to Nodule Infection and Nitrogen-Fixing Ability in the Legume Lotus japonicus. Plant Physiol. 2012;160:2137–2154. doi: 10.1104/pp.112.204883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson KEP, Firoozabady E. Transformation of floriculture crops. Sci. Hortic. (Amsterdam). 1993;55:83–99. doi: 10.1016/0304-4238(93)90026-M. [DOI] [Google Scholar]
- 25.LingFei X, FengWang M, Dong L. Plant regeneration from in vitro cultured leaves of Lanzhou lily (Lilium davidii var. unicolor) Sci. Hortic. (Amsterdam). 2009;119:458–461. doi: 10.1016/j.scienta.2008.08.026. [DOI] [Google Scholar]
- 26.Zhang X, Wang H, Tzi BN. Isolation and characterization of a novel trypsin inhibitor from fresh lily bulbs. Planta Med. 2008;74:546–550. doi: 10.1055/s-2008-1074502. [DOI] [PubMed] [Google Scholar]
- 27.Kwon OK, et al. Anti-inflammatory effects of methanol extracts of the root of Lilium lancifolium on LPS-stimulated Raw264.7 cells. J. Ethnopharmacol. 2010;130:28–34. doi: 10.1016/j.jep.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Lee E, Yun N, Jang YP, Kim J. Lilium lancifolium Thunb. extract attenuates pulmonary inflammation and air space enlargement in a cigarette smoke-exposed mouse model. J. Ethnopharmacol. 2013;149:148–156. doi: 10.1016/j.jep.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Zhao L, Liu H, Cai G, Xia M. Assessment of the genetic diversity and genetic relationships of Lilium in China using ISSR markers. Biochem. Syst. Ecol. 2014;55:184–189. doi: 10.1016/j.bse.2014.03.024. [DOI] [Google Scholar]
- 30.Yamagishi M. How genes paint lily flowers: Regulation of colouration and pigmentation patterning. Sci. Hortic. (Amsterdam). 2013;163:27–36. doi: 10.1016/j.scienta.2013.07.024. [DOI] [Google Scholar]
- 31.Wang J, et al. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genomics. 2014;15:203. doi: 10.1186/1471-2164-15-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, et al. De novo assembly and characterization of stress transcriptome and regulatory networks under temperature, salt and hormone stresses in Lilium lancifolium. Mol. Biol. Rep. 2014;41:8231–8245. doi: 10.1007/s11033-014-3725-1. [DOI] [PubMed] [Google Scholar]
- 33.Xu X, et al. High-Throughput Sequencing and Degradome Analysis Identify miRNAs and Their Targets Involved in Fruit Senescence of Fragaria ananassa. PLoS One. 2013;8:e70959. doi: 10.1371/journal.pone.0070959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33 (2005). [DOI] [PMC free article] [PubMed]
- 36.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA Targets Identified by Sequencing of the Arabidopsis Degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu S, et al. Identification and differential regulation of microRNAs in response to methyl jasmonate treatment in Lycoris aurea by deep sequencing. BMC Genomics. 2016;17:789. doi: 10.1186/s12864-016-2645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan L, Wang X, Jin J, Yu X, Hu J. Bioinformatic Identification and Expression Analysis of Nelumbo nucifera MicroRNA and Their Targets. Appl. Plant Sci. 2015;3:1500046. doi: 10.3732/apps.1500046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao D, Gong S, Hao Z, Tao J. Identification of miRNAs responsive to Botrytis cinerea in herbaceous peony (Paeonia lactiflora Pall.) by high-throughput sequencing. Genes (Basel) 2015;6:918–934. doi: 10.3390/genes6030918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, et al. Characterization and comparative profiling of the small RNA transcriptomes in two phases of flowering in Cymbidium ensifolium. BMC Genomics. 2015;16:622. doi: 10.1186/s12864-015-1764-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, et al. Identification and characterization of MicroRNAs in Ginkgo biloba var. epiphylla mak. PLoS One. 2015;10:1–18. doi: 10.1371/journal.pone.0127184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, et al. Identification and profiling of novel and conserved microRNAs during the flower opening process in Prunus mume via deep sequencing. Mol. Genet. Genomics. 2014;289:169–183. doi: 10.1007/s00438-013-0800-6. [DOI] [PubMed] [Google Scholar]
- 43.Wang X-J, Reyes JL, Chua N-H, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunkar, R., Zhou, X., Zheng, Y., Zhang, W. & Zhu, J.K. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant biol. 8, 25 (2008). [DOI] [PMC free article] [PubMed]
- 45.Lv S, et al. Identification and characterization of microRNAs from barley (hordeum vulgare L.) by high-throughput sequencing. Int. J. Mol. Sci. 2012;13:2973–2984. doi: 10.3390/ijms13032973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan H, et al. Genome-Wide Analysis of MicroRNA Responses to the Phytohormone Abscisic Acid in Populus euphratica. Front. Plant Sci. 2016;7:1–18. doi: 10.3389/fpls.2016.01184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morin, R. D. et al. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. 571–584, 10.1101/gr.6897308.1 (2008). [DOI] [PMC free article] [PubMed]
- 49.Wang L, Liu H, Li D, Chen H. Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genomics. 2011;12:154. doi: 10.1186/1471-2164-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barakat A, Wall PK, DiLoreto S, dePamphilis CW, Carlson JE. Conservation and divergence of microRNAs in Populus. BMC Genomics. 2007;8:481. doi: 10.1186/1471-2164-8-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han L, et al. Identification and Characterization of Erysiphe necator-Responsive MicroRNAs in Chinese Wild Vitis pseudoreticulata by High-Throughput Sequencing. Front. Plant Sci. 2016;7:1–14. doi: 10.3389/fpls.2016.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F, Johnson NR, Coruh C, Axtell MJ. Genome-wide analysis of single non-Templated nucleotides in plant endogenous siRNAs and miRNAs. Nucleic Acids Res. 2016;44:7395–7405. doi: 10.1093/nar/gkw457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chellappan P, et al. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 2010;38:6883–6894. doi: 10.1093/nar/gkq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi S, et al. Sorting of Small RNAs into Arabidopsis Argonaute Complexes Is Directed by the 5′ Terminal Nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeong D-H, et al. Massive Analysis of Rice Small RNAs: Mechanistic Implications of Regulated MicroRNAs and Variants for Differential Target RNA Cleavage. Plant Cell. 2011;23:4185–4207. doi: 10.1105/tpc.111.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X. microRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones-Rhoades MW, Bartel DP. Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 58.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beauclair L, Yu A, Bouché N. MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010;62:454–462. doi: 10.1111/j.1365-313X.2010.04162.x. [DOI] [PubMed] [Google Scholar]
- 60.Lu S, Yang C, Chiang VL. Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa. J. Integr. Plant Biol. 2011;53:879–891. doi: 10.1111/j.1744-7909.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang M, Li C, Lu S. Origin and evolution of MIR1444 genes in Salicaceae. Sci. Rep. 2017;7:39740. doi: 10.1038/srep39740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li C, Li D, Li J, Shao F, Lu S. Characterization of the polyphenol oxidase gene family reveals a novel microRNA involved in posttranscriptional regulation of PPOs in Salvia miltiorrhiza. Sci. Rep. 2017;7:44622. doi: 10.1038/srep44622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gómez-Gómez L, Boller T. FLS2: An LRR Receptor-like Kinase Involved in the Perception of the Bacterial Elicitor Flagellin in Arabidopsis. Mol. Cell. 2000;5:1003–1011. doi: 10.1016/S1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 64.Wang D, et al. BKI1 Regulates Plant Architecture through Coordinated Inhibition of the Brassinosteroid and ERECTA Signaling Pathways in Arabidopsis. Mol. Plant. 2017;10:297–308. doi: 10.1016/j.molp.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Y, Liu X, Xu Z, Yang H, Li J. Characterization and enzymatic properties of protein kinase ACR4 from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2017;489:270–274. doi: 10.1016/j.bbrc.2017.05.163. [DOI] [PubMed] [Google Scholar]
- 66.Ohad N, et al. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, et al. PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 2017;267:27–37. doi: 10.1093/jxb/erx360. [DOI] [PubMed] [Google Scholar]
- 68.Naumann U, et al. Genetic evidence that DNA methyltransferase DRM2 has a direct catalytic role in RNA-directed DNA methylation in Arabidopsis thaliana. Genetics. 2011;187:977–979. doi: 10.1534/genetics.110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qüesta JI, Fina JP, Casati P. DDM1 and ROS1 have a role in UV-B induced- and oxidative DNA damage in A. thaliana. Front. Plant Sci. 2013;4:1–12. doi: 10.3389/fpls.2013.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, F. et al. The F-box protein ZYGO1 mediates bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis. Plant Cell29, tpc.00287.2017 (2017). [DOI] [PMC free article] [PubMed]
- 71.De Jong M, Wolters-Arts M, García-Martínez JL, Mariani C, Vriezen WH. The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J. Exp. Bot. 2011;62:617–626. doi: 10.1093/jxb/erq293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou XY, Song L, Xue HW. Brassinosteroids regulate the differential growth of arabidopsis hypocotyls through auxin signaling components IAA19 and ARF7. Mol. Plant. 2013;6:887–904. doi: 10.1093/mp/sss123. [DOI] [PubMed] [Google Scholar]
- 73.Wang F, et al. High-throughput sequencing discovery of conserved and novel MicroRNAs in Chinese cabbage (Brassica rapa L. ssp. pekinensis) Mol. Genet. Genomics. 2012;287:555–563. doi: 10.1007/s00438-012-0699-3. [DOI] [PubMed] [Google Scholar]
- 74.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Z., Jiang, L., Wang, J., Gu, P. & Chen, M. MTide: an integrated tool for the identification of miRNA—target interaction in plants. Bioinformatics 1–2, 10.1093/bioinformatics/btu633 (2014). [DOI] [PubMed]
- 76.Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. Evidence that miRNAs are different from other RNAs. Cell. Mol. Life Sci. 2006;63:246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fahlgren, N. et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One2 (2007). [DOI] [PMC free article] [PubMed]
- 78.Dai X, Zhao PX. PsRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011;39:155–159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 80.Jia, L. et al. Identification of the conserved and novel miRNAs in mulberry by high-throughput sequencing. PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 81.German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- 82.Chiang C-P, et al. Identification of Ice Plant (Mesembryanthemum crystallinum L.) MicroRNAs Using RNA-Seq and Their Putative Roles in High Salinity Responses in Seedlings. Front. Plant Sci. 2016;7:1–18. doi: 10.3389/fpls.2016.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data (Accession Number: SRA633909) in the study can be obtained from SRA database.