Abstract
The transcriptome and proteome encode distinct information that is important for characterizing heterogeneous biological systems. We demonstrate a method to simultaneously characterize the transcriptomes and proteomes of single cells at high throughput using aptamer probes and droplet-based single cell sequencing. With our method, we differentiate distinct cell types based on aptamer surface binding and gene expression patterns. Aptamers provide advantages over antibodies for single cell protein characterization, including rapid, in vitro, and high-purity generation via SELEX, and the ability to amplify and detect them with PCR and sequencing.
Introduction
Cellular differentiation restricts the genetic programs that cells may execute, endowing distinct functions and phenotypes1,2. This enables important abilities, like the generation of tissues and organs; however, dysregulation of this system can lead to diseases, like cancer3. On the microscopic level, important biological structures comprise heterogeneous ensembles of cells working in coordination4–7. The immune system, for example, uses multiple cell types to elicit a response to exogenous threats, prevent autoimmunity, and establish long-term memory8. In cancer, heterogeneity occurs in advanced malignancies and imposes a significant barrier to cure by often ensuring that a fraction of cancer cells resist treatment9–11. For heterogeneous systems, mixed, multicell measurements do not allow resolution of different cells into their functional groups. High throughput single cell transcriptome sequencing12–14 is an effective tool for deconvoluting heterogeneity because it provides ample information to identify cell type15 and infer cell state and function16. Moreover, it leverages the capacity of modern sequencing to analyze tens-of-thousands of cells per experiment, allowing analysis of populations17–19. Nevertheless, gene expression is dynamic and can change due to biologically important or trivial events, making data interpretation challenging. Indeed, there is often poor correlation between transcript count and protein abundance, particularly when measured in single cells20,21.
As the extreme boundary of cells, the membrane carries a molecular fingerprint useful for identifying cell type and function. This information is often complementary to gene expression data, being encoded in the proteome and, thus, containing features not otherwise observable, like post-translational protein modifications. Consequently, for cell type discrimination, surface profiling with antibodies and fluorescence-activated cell sorting (FACS) is the gold standard to classify cells into their myriad types via well-characterized biomarkers22,23. However, the approach requires that each antibody be labeled with a unique fluorophore, limiting multiplexing to tens of antibodies. By swapping fluorophores with mass tags and using a mass spectrometer for the readout, over a hundred antibodies can be used24,25, although this is still far short of the tens-of-thousands of genes, splice-forms, and post-translational modifications actively used by organisms and available for characterization by antibodies. Abseq replaces the mass tags with nucleic acid sequence tags, using droplet-based single cell sequencing for the readout26. Because a sequence tag is encoded by its full nucleobase set, an astronomical number of sequence combinations are available for unique antibody labeling, shattering the multiplexing barrier. Moreover, the microfluidic approaches used to sequence single cell mRNA can be applied to the tags, allowing simultaneous surface and transcriptome profiling of single cells at high throughput27,28. While this provides exciting opportunities for characterizing cells with paired gene expression and protein data, it requires access to high-affinity antibodies. Effective antibodies are available for common targets, but uncommon ones require custom generation, an involved process necessitating antigen purification29,30. Antigen purification can be costly and labor intensive, and may not be possible depending on the native structure of the antigen31,32. Once obtained, the antibodies must be conjugated to sequence tags, an additional step that can impact affinity. To enable simple and effective single cell surface profiling, an optimal approach would obviate the need for custom antibody generation and tag conjugation.
Here, we present Apt-seq, an approach to simultaneously profile the surfaces and transcriptomes of single cells using aptamers and single cell sequencing. Like antibodies, aptamers are affinity probes capable of specific binding to target epitopes33–36, and aptamer binding can be multiplexed. Unlike antibodies, aptamers are nucleic acids in which the nucleobase sequence provides an intrinsic tag that can be read out via DNA sequencing, obviating the need for additional tag conjugation. Moreover, specific, high-affinity aptamers can be readily and inexpensively obtained with in vitro systematic evolution of ligands by exponential enrichment (SELEX)34,36,37. SELEX can be applied directly to living cells, avoiding antigen purification, and shortening the process from months to weeks38–40. This simplifies affinity reagent generation and enables new surface characterization only accessible to aptamers41. We demonstrate Apt-seq by using it to discriminate between cells based on aptamer binding and differences in gene expression.
Results
Aptamers allow single cell surface profiling via droplet barcoding and sequencing
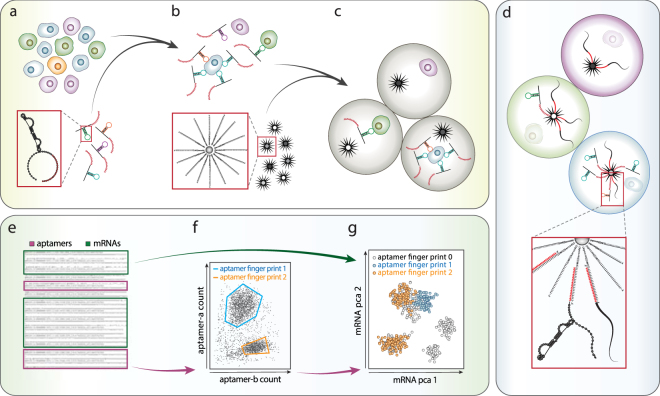
Aptamers are nucleic acids that adopt a three-dimensional fold and bind specifically to protein epitopes and small molecules34,36. Like antibodies, they can be used in combination for multiplexed characterization42, while being easily identified via nucleic acid sequencing. To allow simultaneous sequencing of cell mRNA and aptamers, we polyadenylate the aptamers to mimic the structure of mRNA; this allows both to be captured and sequenced using identical poly-thymine primers (Fig. 1a). To label the cells with aptamers, the mixed aptamer library is incubated with a cell suspension, and unbound aptamers washed away (Fig. 1b). To barcode the cells, we employ Drop-seq, a high throughput microfluidic approach13, although other barcoding methods can also be used12,43–45. In Drop-seq, cells are isolated in droplets with barcoded beads and lysis buffer (Fig. 1c)13. Upon lysis, aptamers and mRNA hybridize to poly-thymine barcode sequences on the beads (Fig. 1d), followed by demulsification, washing, and nucleic acid amplification12,46–48. Amplification conjugates a unique barcode sequence to all aptamers and transcripts of a single cell, allowing material for many cells to be pooled, sequenced, and computationally deconvoluted by barcode. This provides, for every cell, paired aptamer and transcript reads (Fig. 1e) that are separated in silico (Fig. 1f,g).
Figure 1.
Principle of the Apt-seq workflow. (a) A heterogeneous cell sample is incubated with a diverse aptamer library containing a poly-A sequence on its 3′-end. (b) Cells expressing epitopes of interest are decorated by the corresponding aptamers in the library and non-binding aptamers are washed away. (c) Single cells of the washed cell suspension are co-encapsulated with beads carrying a unique DNA barcode in a microfluidic device. (d) Each droplet contains lysis solution to lyse cells. Aptamers and mRNA molecules can hybridize with the barcoding beads by means of their poly-A sequence. Using the barcode bead as a primer in reverse transcription and DNA polymerase reactions, the droplet-specific unique barcode is fused to the mRNA and aptamer, providing a cell specific identifier. (e) Pooling all beads after barcode fusion, sequencing their content in parallel, and deconvoluting aptamers and mRNAs, allows evaluation of epitope profiles in single cells (f). (g) Since the cell-specific barcode is shared between aptamers and transcripts, the epitope data can be combined with the single cell transcriptome for further interdependent analysis.
Polyadenylation does not impair aptamer function
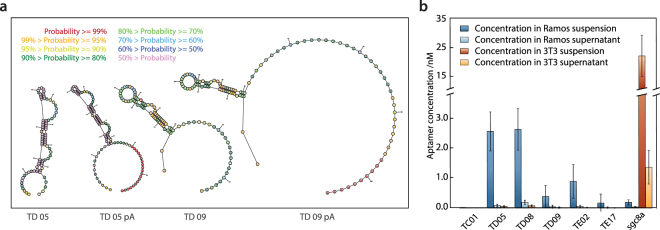
For Apt-seq to be effective, the poly-adenylation required for paired transcriptome sequencing must not perturb aptamer binding49. To confirm this, we construct a library of five aptamers, TC01, TD05, TD08, TD09, and TE02, reported to bind Ramos cells with Kd from 0.8 nM to 74.7 nM50. We also include TE17, sgc3b, and sgc8a aptamers that do not bind Ramos cells42,50,51. TD05, sgc3b, and sgc8a have reported protein targets, the membrane bound IgM, L-selectin, and PKT7, respectively52–54. To assess the impact of the poly-A tail on aptamer fold, we use RNAstructure55, a secondary structure prediction algorithm, and predict the same fold for the aptamers with and without poly-A tail (Fig. 2a). To assess whether the tails interfere with binding, we synthesize all eight polyadenylated aptamers and apply them to Ramos and control 3T3 cells. The aptamers are incubated at equal molar concentration with either cell line, followed by five wash cycles and concentration estimation in the final wash supernatant and final cell suspension by qPCR. In agreement with previous studies, TD05, TD08, and TE02 are highly enriched in Ramos cell suspensions, while TD09 is moderately enriched. In contrast, TC01, TE17, and sgc8a are not enriched in Ramos cells, as expected (Fig. 2b) (Supplementary Fig. 1). Sgc3b remains below the detection limit for either cell line. Notably, although sgc8a is a reported binder of human T-cells42, it enriches in mouse 3T3 cells. However, a previous study showed that both sgc8 and a PKT7 binding antibody can interact with other cell lines presumably devoid of PKT756. We conclude that poly-adenylating the aptamers does not affect fold or binding and that all except TC01 perform as previously reported in our hands.
Figure 2.
Influence of the 3′-poly-A tail on aptamer structure and function. (a) Predicted secondary structure of the aptamers TD05 and TD09 with and without poly-A tail. (b) The functionality of the aptamers TC01, TD05, TD08, TD09, TE02, TE17 and sgc8a, all modified with a 3′-poly-A tail is evaluated based on their ability to bind to Ramos or 3T3 cells. The concentration of the aptamers is estimated in the supernatant and cellular fraction after five wash cycles by qPCR. Each bar represents the mean of a technical triplicate and uncertainty is given as one sample standard deviation.
Apt-seq provides independent information from RNAseq for inferring cell type
Inferring cell type from RNAseq data can be challenging due to the dynamic, complex, and multidimensional nature of gene expression. An advantage of Apt-seq is that it allows independent confirmation of cell type by aptamer binding. This can be used to support inferences from transcriptome data, or provide additional information for differentiating between related cells and states. To illustrate these benefits, we perform combined aptamer and transcriptome sequencing of suspended Ramos and 3T3 cells. After incubation and washing of the aptamers, we barcode the cells with Drop-seq. Drop-seq uses template switching to generate defined 3′-ends on cDNA, enabling subsequent PCR amplification. After amplification, cDNA is tagmented to generate ~500 bp fragments containing necessary sequencing handles. Aptamers lack the 5′-cap of mRNA and, thus, template switching is non-processive, preventing addition of the handles by this route. However, our aptamers are constructed with known flanking sequences, which we use for amplification and handle attachment. Additionally, the aptamers are short (below 200 bp), so that amplicons can be sequenced without tagmentation (Methods, Supplementary Tables 1 and 2). These simplifications allow efficient, joint processing of aptamers and mRNA in the same barcoding reaction.
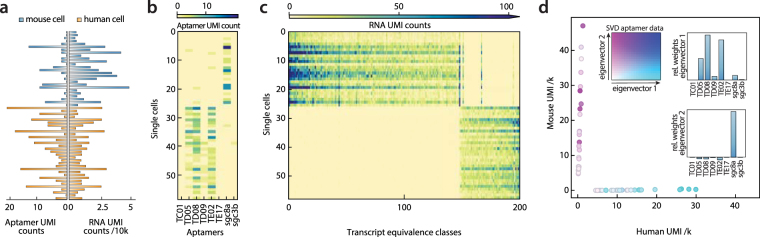
After barcoding, the nucleic acids of many cells are pooled and sequenced. We aim for about 200 cells and perform limited Illumina MiSeq sequencing collecting ~12 million paired-end reads. We computationally group reads to single cells using barcodes, and then to single molecules using unique molecular identifiers (UMIs). After processing, we obtain high-quality transcriptome and aptamer data for 58 cells, with >4500 unique reads per cell. More cells can be obtained by sequencing deeper and collecting more beads13. As expected, most reads belong to the transcriptome, and a smaller fraction to surface-bound aptamers (Fig. 3a). The aptamer profiles segregate into two groups, with TD08 and TE02, and to a lesser degree TD05 and TD09. The aptamer sgc8a is predominantly anticorrelated with this group. We obtain few reads for TC01, TE17, and sgc3b (Fig. 3b), consistent with our multicell qPCR results (Fig. 2b). Based on the qPCR results and because TD05 is a binder of the immunoglobulin heavy chain, we expect the first rectangular block to represent Ramos and the second 3T3 cells. Because these cells are from different species, cell type can be inferred by direct sequence analysis of cDNA. To verify our aptamer results, we thus evaluate the transcript reads of each cell. We order the transcriptome data using the same y-axis as the aptamers and again observe a clear block segregation, indicating that the gene expression of the two cell types is differentiated as expected. Based on transcript sequences, we confirm that the lower block corresponds to the human Ramos and the upper block to the mouse 3T3 cells (Fig. 3c).
Figure 3.
Mixed human-mouse two cell type experiment. (a) The number of unique aptamer reads per cell is drawn on the left. On the opposite side, all unique mRNA reads are displayed. The cells are ordered from bottom to top according to the number of assigned human mRNA reads divided by the total number of assigned mRNAs in decreasing order. (b) Two-dimensional histogram of counts per aptamer and cell. The order of the cells is the same as in (a). (c) Two-dimensional histogram of the transcript counts for the 200 most variable transcript equivalence classes. The maximum read number is truncated at 100. The order of the cells is the same as in (a,b). (d) Barnyard plot of the two cell transcriptome data. The first two principal components of the cell versus aptamer read matrix are superimposed in color space. Color code is given in the insert. The relative contribution of each aptamer to the first two principal components is given.
To confirm the relationship between expected aptamer and gene expression data, we construct a barnyard plot, displaying each cell as a 2D coordinate corresponding to the number of human and mouse transcripts identified within its barcode cluster (Fig. 3d). We extract the principal components of the aptamer read per cell matrix by calculating a singular value decomposition (SVD) (Fig. 3d, upper-left). This yields two eigenvector principle components, describing how the aptamers tend to correlate with each other. In agreement with the observed block structure of the aptamer profiles, we obtain an all positive first eigenvector with major contribution of the aptamers TD05, TD08, and TE02 (Fig. 3d, upper right) and a second eigenvector consisting primarily of sgc8a (Fig. 3d, lower-right). These eigenvectors are strongly anticorrelated, indicating that the aptamers cluster cleanly based on cell type. We use these eigenvectors to define a two-parameter coloring scheme, coloring each data point corresponding to a cell in the coordinate space according to its amplitudes along the two eigenvectors. Cells containing mostly human transcripts have amplitudes primarily along the first eigenvector (blue, Fig. 3c upper-right), while cells containing mostly mouse transcripts have amplitudes primarily along the second eigenvector (purple, Fig. 3d, lower-right). These results demonstrate that transcriptional signatures cluster with known patterns of aptamer binding and, more broadly, that aptamers can be used to label specific cell types in a format that can be read out with mRNA sequencing data.
Discussion
While single cell mRNA profiling provides a valuable snapshot into the inner workings of cells, it is just one of many characterizations of the cellular space. Inaccessible from transcriptome data, the proteome contains traces of post-translational regulatory events and exhibits different temporal dynamics57,58. Tapping into both domains, Apt-seq provides a strategy for highly multiplexed, single cell surface profiling simultaneous with the transcriptome. Because Apt-seq uses polyadenylated aptamer probes, it is compatible with most common single cell transcriptome sequencing methods, including Drop-seq, InDrops, and their commercial variants.
Apt-seq mirrors Abseq, and its successors cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) and RNA expression and protein sequencing (REAP-seq), in that it allows combined surface and transcriptome profiling of tens-of-thousands of single cells at high throughput26–28. However, aptamers provide practical and functional advantages over antibodies. In practical terms, aptamers are faster and cheaper to generate against many novel targets than antibodies. The in vitro SELEX process can be performed directly on living cells, obviating the need for antigen purification, which is not possible for many targets. Thus, Abseq may be preferred when the marker is best detected by a readily-available antibody, while Apt-seq may be superior for novel targets. Additionally, while antibodies require tag conjugation, aptamers can be used directly, since they are already sequenceable nucleic acids. In addition, patents on the original SELEX method have expired and new protocols have emerged that should expand the library of high-quality, specific aptamers37,59–61. Moreover, aptamers can be synthesized with higher purity and reliability than antibodies through established chemical synthesis pipelines; this may allow aptamers to overcome the poor reproducibility that has plagued antibodies, and potentially achieve higher experimental reliability.
Apt-seq provides new capabilities for high throughput single cell characterization. In addition to scaling numbers of cells sequenced per sample, there is an increasing interest in scaling the number of samples sequenced in total. Ideally, many samples would be pooled and sequenced on a single run, but the need to index each one necessitates microfluidic barcoding of every sample, an expensive and labor-intensive process. Apt-seq can address this by labeling samples with unique surface aptamers that can be used independent of transcriptome data to batch samples together in one microfluidic barcoding run. Since after cell washing the free aptamer concentration is orders of magnitude below their dissociation constant, there should be minimal cross contamination between samples.
Aptamers and antibodies can bind internal epitopes of cells using fixation and permeabilization protocols; however, such binding may be better with the smaller aptamers, having a hydrodynamic diameter of ~2 nm compared to ~15 nm for antibodies41,62. Indeed, many aptamers are readily internalized and staining of intracellular compartments can be more effective than with antibodies63–65. This will be important for combining intracellular proteomic and transcriptomic readouts66. Aptamers also enable measurement opportunities not possible with antibodies. For example, a raw aptamer library produced by cell-SELEX can be directly applied to cells, allowing different cell types to be inferred based on aptamer spectrum, rather than assembling an aptamer library from known binders. This should allow differentiation between cells even when distinct biomarkers are not known a priori. Additionally, multidentate, bispecific, or multipiece aptamers can be constructed by combining binding sequences in the same aptamer67,68, allowing assessment of epitope proximity in a way currently accomplished with fluorescence or proximity ligation assays, but in a format that can be highly multiplexed and performed simultaneously with cell surface and transcriptome profiling. Furthermore, riboswitch-like aptamers have a unique ability to bind small molecules and, thus, Apt-seq opens new possibilities for adding cellular metabolite characterization to simultaneous protein and nucleic acid measurements in single cells69.
Methods
Aptamer structure prediction
The aptamer structure was predicted by the RNAstructure web-server (https://rna.urmc.rochester.edu/RNAstructureWeb/) with default parameters55.
Cell cultures
Ramos and 3T3 cells were cultured at 37 °C in RPMI-1640 medium supplemented with antibiotics and 5% fetal bovine serum (FBS) in the presence of 5% CO2.
Aptamer staining
Aptamers (from IDT) were folded as described before50. Briefly, they were diluted to 0.5 μM in aptamer folding buffer (F) (1× PBS, 5 mM MgCl2, 4.5 gl−1 glucose) and heated to 95 °C for five minutes then cooled on ice for 10 min. The folded aptamers were pooled at a concentration of 62.5 nM each as stock solution.
3T3 and Ramos cells were resuspended in aptamer binding buffer (B) (buffer F supplemented with 1 mg/ml BSA and 0.1 mg/ml salmon sperm DNA). For the qPCR experiments either 3T3 or Ramos cells were diluted to about 6 × 105 cells/ml and incubated with 31.25 nM of each aptamer. Cells were washed five times by centrifugation and resuspension cycles. For the single cell experiment 106 Ramos cells where mixed with 3*105 3T3 cells and 31.25 nM of each aptamer in 1 ml buffer B. The cell suspension was incubated for 30 min on ice and washed in ice cold buffer B for five centrifugation and resuspension cycles.
Quantitative PCR
For each qPCR replicate either a suspension of about 4200 aptamer-stained cells or, in controls, the corresponding volume of supernatant was loaded. Aptamer specific primers (from IDT) were used to amplify individual sequences (Supplementary Table 2). Signal was detected on a QuantStudio5 (Applied Biosystems) qPCR machine with SYBR green as reporter. A control experiment was performed to assess background priming on non-target aptamers and was found to be below detection limit. Amplification standard curves were measured for all aptamer primer pairs in triplicate. The model C(n) = C0bn, where C(n) corresponds to the concentration after n PCR cycles and C0 to the start concentration, was log transformed and fit by linear regression to obtain the amplification rate b and the detection concentration C(nCt), which is reached at the cycle threshold (Ct). Both calculated constants were used to estimate the aptamer concentration in the experiment. Uncertainty in the constants was propagated and final uncertainty of the concentration estimates is presented as one sample standard deviation with applied Bessel’s correction.
Single cell experiment
The aptamer-stained cell suspension was diluted in ice cold PBS containing 0.1% BSA to a final concentration of 1.06*105 cells/ml.
Single cell experiments were performed as described by13 (online protocol:70) with the following modifications. About 2000 beads were used for further processing, corresponding to about 100–200 cells. After reverse transcription with Maxima H Minus Reverse Transcriptase (Thermo Fisher), which produces barcoded cDNA-mRNA hybrids and barcoded double-stranded aptamers, we amplified both libraries together by 13 cycles of PCR. DropSeq uses template switching to introduce the sequence ACTCTGCGTTGATACCACTGCTT at the 3′-end of the cDNA which allows use of a single primer (smart PCR) (Supplementary Table 2) to amplify the complete barcoded construct. Since template switching is much more efficient on 5′-capped mRNA, it only rarely happens on aptamers. Therefore, we use a specific reverse primer for each different aptamer flanking region (Supplementary Table 1), which we add to the PCR mix at a ratio of 1:20 compared to the smart PCR primer. Next DropSeq uses tagmentation (Nextera) to trim the transcripts to a mean length of about 500 bp and to introduce the sequence overhang GTCTCGTGGGCTCGG. Since the length of the aptamers is significantly shorter, tagmentation is not expected to act on them. To enable downstream joint processing of aptamers and transcripts we introduced the tagmentation overhang as part of the aforementioned specific primers. We processed the sample further as described13. The cDNA library was sequenced on a MiSeq device (Illumina) with a MiSeq Reagent Kit v3 (Illumina) in paired-end mode.
Sequencing and data analysis
The python scripts for the evaluation of InDrops experiments71 was adjusted to be applicable to the barcoding scheme of the DropSeq experiment. Without attempting any error correction, in a first pass all cellular barcodes with a count above 2000 where retained, which yielded 210 distinct barcodes. The corresponding sequences, containing transcripts and aptamers, were evaluated with Kallisto in the UMI-aware mode to generate a pseudo-alignment and to map transcripts to equivalence classes72,73. To reduce the influence of stochastic fluctuations in the data, cells were only included for further analysis if they yielded a minimum of 4500 unique transcript reads; 58 cells passed this filter. For display purpose, the 200 equivalence classes with the highest variance were selected (Fig. 3c). A barnyard plot was generated based on equivalence classes that could be unambiguously assigned to either mouse or human only (Fig. 3d). Aptamer sequences were identified separately by the program cutadabt74. For each aptamer three overlapping 20mers were generated and each of these fragments was used as a cutadabt query and tested against all sequences. If a sequence contained a consecutive 20mer that matched the query with at most one base mismatch, the sequence was considered a read of that aptamer. The identified aptamer sequences were collapsed based on their UMI. SVD was calculated based on the mean-centered normalized cells versus aptamers count matrix. The color of the x-coordinate in the barnyard plot corresponds to the amplitude of the first eigenvector (cyan) and the y-coordinate to the inverse amplitude of the second eigenvector (magenta) (Fig. 3d).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Electronic supplementary material
Acknowledgements
We thank John Haliburton for helping with Drop-seq setup, Anna Sellas of the Chan Zuckerberg Biohub for assisting with the library sequencing and Sarah Pyle for helping with the figures. This work was supported by the Chan Zuckerberg Biohub, the National Science Foundation CAREER Award (Grant Number DBI-1253293); the National Institutes of Health (NIH) (Grant Numbers R01-EB019453-01, R01-HG008978 and DP2-AR068129-01); C.L.D. is supported by a Swiss National Science Foundation fellowship (Grant Number 175086).
Author Contributions
A.R.A. conceived the project. C.L.D., L.L. and M.F.S. designed and carried out the experiments. C.L.D. evaluated the data. A.R.A., C.L.D. wrote and all authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21153-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes & Development. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 2.Murry CE, Keller G. Differentiation of Embryonic Stem Cells to Clinically Relevant Populations: Lessons from Embryonic Development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Friedmann‐Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO reports. 2014;15:244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nature Reviews Genetics. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- 5.Gerlinger M, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. The New England Journal of Mecdicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vickaryous MK, Hall BK. Human cell type diversity, evolution, development, and classification with special reference to cells derived from the neural crest. Biological Reviews. 2006;81:425–455. doi: 10.1017/S1464793106007068. [DOI] [PubMed] [Google Scholar]
- 8.Satija R, Shalek AK. Heterogeneity in immune responses: from populations to single cells. Trends in Immunology. 2014;35:219–229. doi: 10.1016/j.it.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nature Biotechnology. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 10.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein AM, et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Reviews Genetics. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 15.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 16.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nature Biotechnology. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg, A. B. et al. Scaling single cell transcriptomics through split pool barcoding. bioRxiv. 10.1101/105163 (2017)
- 18.Cao J, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habib N, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Meth. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elowitz MB, Levine AJ, Siggia ED. Stochastic Gene Expression in a Single Cell. Science. 2002;297:1183–1187. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 22.Robinson JP, Roederer M. Flow cytometry strikes gold. Science. 2015;350:739–740. doi: 10.1126/science.aad6770. [DOI] [PubMed] [Google Scholar]
- 23.Fulwyler MJ. Electronic separation of biological cells by volume. Science. 1965;150:910–911. doi: 10.1126/science.150.3698.910. [DOI] [PubMed] [Google Scholar]
- 24.Bandura DR, et al. Mass Cytometry: A Novel Technique for Real-Time Single Cell Multi-Target Immunoassay Based on Inductively Coupled Plasma Time of Flight Mass Spectrometry. Analytical Chemistry. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 25.Bendall SC, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:687–695. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahi P, Kim SC, Haliburton JR, Gartner ZJ, Abate AR. Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Scientific Reports. 2017;7:44447. doi: 10.1038/srep44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson VM, et al. Multiplexed quantification of proteins and transcripts in single cells. Nature Biotechnology. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 28.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seeber S, et al. A robust high throughput platform to generate functional recombinant monoclonal antibodies using rabbit B cells from peripheral blood. PLoS ONE. 2014;9:1–14. doi: 10.1371/journal.pone.0086184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith K, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nature Protocols. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamakubo T, Kusano-Arai O, Iwanari H. Generation of antibodies against membrane proteins. Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics. 2014;1844:1920–1924. doi: 10.1016/j.bbapap.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Takeda H, et al. Production of monoclonal antibodies against GPCR using cell-free synthesized GPCR antigen and biotinylated liposome-based interaction assay. Scientific Reports. 2015;5:11333. doi: 10.1038/srep11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn MR, Jimenez RM, Chaput JC. Analysis of aptamer discovery and technology. Nature Reviews Chemistry. 2017;1:76. doi: 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- 34.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 35.Keefe AD, Pai S, Ellington A. Aptamers as Therapeutics. Nature Reviews Drug Discovery. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 37.Gotrik MR, et al. Advancements in Aptamer Discovery Technologies. Accounts of Chemical Research. 2016;49:1903–1910. doi: 10.1021/acs.accounts.6b00283. [DOI] [PubMed] [Google Scholar]
- 38.Daniels DA, et al. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proceedings of the National Academy of Sciences. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shangguan D, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. Journal of Proteome Research. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao Z, et al. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry—A European Journal. 2008;14:1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nature Reviews Drug Discovery. 2016;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shangguan D, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11838–43. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gierahn TM, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nature Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nature Communications. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimshony T, et al. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplificatio. Cell Reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome research. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cowperthwaite MC, Ellington AD. Bioinformatic Analysis of the Contribution of Primer Sequences to Aptamer Structures. Journal of Molecular Evolution. 2008;67:95–102. doi: 10.1007/s00239-008-9130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Z, et al. Selection of Aptamers for Molecular Recognition and Characterization of Cancer Cells. Analytical Chemistry. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 51.Rincon J, Prieto J, Patarroyo M. Expression of integrins and other adhesion molecules in Epstein-Barr virus-transformed B lymphoblastoid cells and Burkitt’s lymphoma cells. International journal of cancer. 1992;51:452–8. doi: 10.1002/ijc.2910510319. [DOI] [PubMed] [Google Scholar]
- 52.Bing T, Shangguan D, Wang Y. Facile Discovery of Cell-surface Protein Targets of Cancer Cell Aptamers. Molecular & cellular proteomics. 2015;10:2692–2700. doi: 10.1074/mcp.M115.051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mallikaratchy P, et al. Aptamer Directly Evolved from Live Cells Recognizes Membrane Bound Immunoglobin Heavy Mu Chain in Burkitt’s Lymphoma Cells. Molecular & Cellular Proteomics. 2007;6.12:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Mallikaratchy P, Xiao Z. Optimization and Modifications of Aptamers Selected from Live Cancer Cell Lines. Chembiochem. 2007;2:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 55.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129–129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li N, et al. Technical and Biological Issues Relevant to Cell Typing with Aptamer. Journal of Proteome Research. 2009;8:2438–2448. doi: 10.1021/pr801048z. [DOI] [PubMed] [Google Scholar]
- 57.Waldbauer JR, et al. Transcriptome and Proteome Dynamics of a Light-Dark Synchronized Bacterial Cell Cycle. PLoS ONE. 2012;7:1–13. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nature Biotechnology. 2003;21:255. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 59.Birch CM, et al. Identification of malaria parasite-infected red blood cell surface aptamers by inertial microfluidic SELEX (I-SELEX) Scientific Reports. 2015;5:11347–11347. doi: 10.1038/srep11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, et al. Particle Display: A Quantitative Screening Method for Generating High Affinity Aptamers. Angewandte Chemie. 2014;126:1–6. doi: 10.1002/ange.201310509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, et al. Multiparameter Particle Display (MPPD): A Quantitative Screening Method for the Discovery of Highly Specific Aptamers. Angewandte Chemie—International Edition. 2017;56:744–747. doi: 10.1002/anie.201608880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orava EW, Cicmil N, Gariépy J. Delivering cargoes into cancer cells using DNA aptamers targeting internalized surface portals. Biochimica et Biophysica Acta (BBA)—Biomembranes. 2010;1798:2190–2200. doi: 10.1016/j.bbamem.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 63.De Castro MAG, Hobartner C, Opazo F. Aptamers provide superior stainings of cellular receptors studied under superresolution microscopy. PLoS ONE. 2017;12:1–16. doi: 10.1371/journal.pone.0173050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magalhães MLB, et al. A General RNA Motif for Cellular Transfection. Molecular Therapy. 2012;20:616–624. doi: 10.1038/mt.2011.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ray, P. & White R. R. Cell-SELEX Identifies a “sticky” RNA Aptamer Sequence. Journal of Nucleic Acids. 2017, (2017). [DOI] [PMC free article] [PubMed]
- 66.Baron M, Yanai I. New skin for the old RNA-Seq ceremony: the age of single-cell multi-omics. Genome Biology. 2017;18:159–159. doi: 10.1186/s13059-017-1300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boltz A, et al. Bi-specific aptamers mediating tumor cell lysis. Journal of Biological Chemistry. 2011;286:21896–21905. doi: 10.1074/jbc.M111.238261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mallikaratchy PR, et al. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Research. 2011;39:2458–2469. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKeague, M. & Derosa, M. C. Challenges and opportunities for small molecule aptamer development. Journal of Nucleic Acids. 2012, (2012). [DOI] [PMC free article] [PubMed]
- 70.Macosko, E., Goldman, M. & McCarroll, S. Drop‐Seq Laboratory Protocol, v3.1, at McCarroll Lab, http://mccarrolllab.com/dropseq/ (2015)
- 71.Zilionis R, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nature Protocols. 2016;12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- 72.Bray N, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 73.Ntranos V, et al. Fast and accurate single-cell RNA-Seq analysis by clustering of transcript-compatibility counts. Genome Biology. 2016;17:1–14. doi: 10.1186/s13059-016-0970-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:5–7. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.