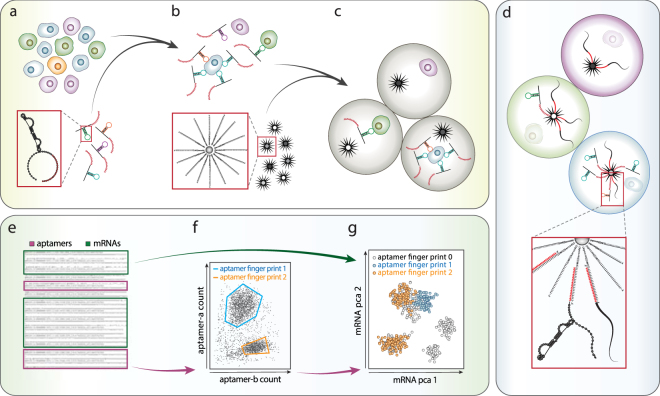
Figure 1.
Principle of the Apt-seq workflow. (a) A heterogeneous cell sample is incubated with a diverse aptamer library containing a poly-A sequence on its 3′-end. (b) Cells expressing epitopes of interest are decorated by the corresponding aptamers in the library and non-binding aptamers are washed away. (c) Single cells of the washed cell suspension are co-encapsulated with beads carrying a unique DNA barcode in a microfluidic device. (d) Each droplet contains lysis solution to lyse cells. Aptamers and mRNA molecules can hybridize with the barcoding beads by means of their poly-A sequence. Using the barcode bead as a primer in reverse transcription and DNA polymerase reactions, the droplet-specific unique barcode is fused to the mRNA and aptamer, providing a cell specific identifier. (e) Pooling all beads after barcode fusion, sequencing their content in parallel, and deconvoluting aptamers and mRNAs, allows evaluation of epitope profiles in single cells (f). (g) Since the cell-specific barcode is shared between aptamers and transcripts, the epitope data can be combined with the single cell transcriptome for further interdependent analysis.