Abstract
Neuroendocrine tumours (NETs) may arise throughout the body and are a highly heterogeneous, relatively rare class of neoplasms difficult to study also for the lack of disease models. Despite this, knowledge on their molecular alterations has expanded in the latest years, also building from genetic syndromes causing their onset. Pancreatic NETs (PanNETs) have been among the most studied, and research so far has outlined a series of recurring features, as inactivation of MEN1, VHL, TSC1/2 genes and hyperactivation of the PI3K/mTOR pathway. Next-generation sequencing has added new information by showing the key role of alternative lengthening of telomeres, driven in a fraction of PanNETs by inactivation of ATRX/DAXX. Despite this accumulation of knowledge, single studies often relied on few cases or were limited to the DNA, RNA, protein or epigenetic level with lack of integrative analysis. The International Cancer Genome Consortium aimed at removing these barriers through a strict process of data and samples collection, to produce whole-genome integrated analyses for many tumour types. The results of this effort on PanNETs have been recently published and, while confirming previous observations provide a first snapshot of how heterogeneous is the combination of genetic alterations that drive this tumour type, yet converging into four pathways whose alteration has been enriched by newly discovered mechanisms. While calling for further integration of genetic and epigenetic analyses, these data allow to reconcile previous findings in a defined frame and may provide clinical research with markers for patients stratification and to guide targeted therapy decisions.
Keywords: neuroendocrinology, pancreas, cancer, molecular biology
Introduction
Neuroendocrine tumours (NETs) are relatively rare, with an incidence of 3–5 cases per 100,000 individuals per year in the United States and Europe, and have a variable behaviour from relatively indolent to highly aggressive (Lloyd et al. 2017). However, a large number of these tumours have a prolonged clinical course, this causing their prevalence to be one of the highest, second only to colorectal cancer according to the National Cancer Institute Surveillance, Epidemiology, and End Results program (Yao et al. 2008, Dasari et al. 2017). This observation, together with the fact that the incidence of these tumours has been steadily increasing in the last 30 years, has risen the interest on a better understanding of this largely heterogeneous group of tumours to guide a better management of patients.
NETs may arise in different sites, and the largest group is constituted by gastroenteropancreatic NETs (GEP-NETs, 61%). Despite having been initially known as cause of the ‘carcinoid syndrome’, most GEP-NETs may be asymptomatic for a long time or present with non-specific symptoms. As a result, a large fraction of patients are diagnosed with advanced or metastatic disease, and the mortality rate is 50% (Scherubl et al. 2013). Due to the lack of disease models, that would be anyhow limited by the large heterogeneity of these tumours, most of the research on their biology has been undertaken by investigating tumour specimens (Capdevila et al. 2017). A small fraction (10%) of NETs arise in the context of familial syndromes, and the genetic alterations linked to those syndromes constituted the first information on the biology of NETs that was also investigated in sporadic tumours. The advent of powerful high-throughput techniques such as next-generation sequencing allowed a deeper, unbiased exploration of tumour specimens, which led to the progressive accumulation of further knowledge on the mechanisms that drive the progression towards malignancy (Scarpa et al. 2015). However, these studies still lacked the coordinated efforts to collect, select and analyse the whole genome of a large number of cases, which has been the flagship of the International Cancer Genome Consortium (ICGC) (International Cancer Genome Consortium2010).
In the ICGC framework, the Italian-Australian initiative has been involved in the analysis of pancreatic adenocarcinoma and rare pancreatic neoplasms. The latter section of the project recently led to publishing the results of whole-genome analysis on 98 pancreatic NETs (PanNETs). This study, while confirming the previous knowledge, adds interesting information and a broader understanding of how mutations, chromosomal alterations and gene expression converge on specific pathways (PI3K/mTOR, DNA damage repair, chromatin modification) more often than previously expected, and with new players (Scarpa et al. 2017). In this focused review, we will summarise our knowledge on PanNETs before (Table 1) the publication of the ICGC group results and will then discuss the contribution of the latter to the building of a more complete picture of the molecular biology of PanNETs. Finally, we will give an overview of the open questions and expected developments after this milestone, especially with regard to targeted therapy (Capdevila et al. 2017, Gajate et al. 2017).
Table 1.
Genes implicated in the tumorigenesis of pancreatic neuroendocrine tumours (PanNETs) before the publication of the International Cancer Genome Consortium data.
| Gene/locus | First information source | Pathway |
|---|---|---|
| MEN1 | Genetic syndrome (multiple endocrine neoplasia type I) | Chromatin remodeling, altered telomeres, PI3K/mTOR DNA double-strand break repair |
| VHL | Genetic syndrome (Von-Hippel Lindau) | PI3K/mTOR |
| NF1 | Genetic syndrome (neurofibromatosis type I) | PI3K/mTOR, Ras/MAPK |
| TSC1/TSC2 | Genetic syndrome (tuberous sclerosis) | PI3K/mTOR |
| PTEN | Next-generation sequencing of PanNETs | PI3K/mTOR |
| ATRX | Next-generation sequencing of PanNETs | Telomeres elongation |
| DAXX | Next-generation sequencing of PanNETs | Telomeres elongation, PI3K/mTOR |
| 1q, 3p, 6q, 10q, 11q | Chromosomal abherrations of PanNETs (LOSS) | – |
| 7p, 9p, 17q, 20q | Chromosomal abherrations of PanNETs (GAIN) | – |
| RASSF1A | Promoter hypermethylation in PanNETs | Ras/MAPK |
| CDKN2A | Promoter hypermethylation in PanNETs | Cell cycle |
| ALU, LINE1 | Promoter hypomethylation in PanNETs | – |
| miR-21 | miRNA differential expression in PanNETs | PI3K/mTOR |
The molecular picture of PanNETs from hereditary syndromes and pre-ICGC studies
Inherited cancer predisposition syndromes account for about one-tenth of diagnosed NETs. The genetic study of syndromic subjects and their families provided the first knowledge of pivotal genes that, when altered in the germline, predispose to endocrine tumour onset. The MEN1 gene, identified in patients suffering from multiple endocrine neoplasia type I, has been proven to be altered also in a large fraction (44%) of sporadic PanNETs (Corbo et al. 2010, Jiao et al. 2011) and in a smaller but significant group of pulmonary carcinoids, where it was linked to poorer prognosis (Swarts et al. 2014). The gene product, menin, has several activities linked to its ability to modulate histones methylation, that may be summarised in three broader functions: (i) negative regulation of the cell cycle via increased expression of CDKN2C/CDKN1B, (ii) inhibition of the PI3K/mTOR signalling pathway via regulation of AKT1 cellular localisation and (iii) promotion of DNA double-strand breaks repair through activation of genes belonging to the homologous recombination (HR) DNA repair machinery like BRCA1 and RAD51. The PI3K/mTOR pathway is also affected by three other genetic syndromes that may result in NETs development: Von-Hippel-Lindau (VHL) disease, neurofibromatosis type I (NF1) and tuberous sclerosis (TS). VHL is caused by inactivating mutations in the namesake gene, which is a negative regulator of the of the hypoxia-induced pro-proliferative HIF1 gene, a downstream effector of the PI3K/mTOR pathway as well; the VHL gene has also been reported to be frequently inactivated by non-mutational mechanisms in up to 25% of sporadic PanNETs (Scarpa et al. 2015). Germline mutation of the NF1 gene is the cause of neurofibromatosis type I, as the gene product neurofibromin is a negative regulator of the Ras/MAPK and PI3K/mTOR signal transduction networks; its involvement in sporadic PanNETs is occasional, while about 40% of periampullary duodenal somatostatinomas associate with NF1 disease. Finally, TS arises by inactivating mutation of either of two genes (TSC1 and TSC2), which act as a complex to inhibit PI3K/mTOR signalling downstream of AKT1. While TS rarely results in PanNETs, recent works have shown that TSC2 downregulation and mutation affect around 35% and 9% of sporadic PanNETs, respectively (Missiaglia et al. 2010, Jiao et al. 2011). The latter data about TSC2 mutation in sporadic PanNET was produced by the first whole-exome study of this neoplasia, including 68 cases (Jiao et al. 2011). This study also confirmed previous low-throughput studies on the frequent mutation of MEN1, and identified PTEN, another gene of the PI3K/mTOR pathway involved in the inhibition of PI3K, as a recurrently (7.3%) mutated tumour suppressor.
Moreover, two novel genes were found frequently mutated that flanked MEN1 in the category of the chromatin remodeling genes implicated in PanNET carcinogenesis: ATRX and DAXX (Jiao et al. 2011). These genes were affected by inactivating mutation in 18% and 25% of PanNETs, with mutations in each other gene showing no overlap. This mutual exclusivity of ATRX/DAXX mutations led to the hypothesis of a physical interaction between the two gene products, that was confirmed by subsequent research; indeed both DAXX and ATRX interact to bind and deposit histone H3.3 on several sites, including the centromere and the telomeres (Lewis et al. 2010). The inactivation of this complex has been linked to chromosomal instability, poorer prognosis and to a phenomenon called alternative lengthening of telomeres (ALT), where telomeres are elongated in a telomerase-independent way. This association has been described specifically in PanNETs and in correlation with higher stage tumours, suggesting that ATRX/DAXX inactivation is a late event of the neoplastic transformation (Heaphy et al. 2011, de Wilde et al. 2012, Marinoni et al. 2014). Mutations in chromatin remodeling (i.e. MEN1, ATRX, DAXX) and HR DNA repair genes either directly involved in DNA repair (BRCA2) or activated upon double-strand break detection and controlling the intersection between cell cycle, DNA repair and apoptosis (CHEK2), support the idea of a lead role for chromosomal and epigenetic alterations in the tumorigenesis of PanNET. Indeed, mutations alone provide a rationale for tumorigenesis only in about 40% of these neoplasms. This means that in the remaining cases, the initiation of the neoplastic process may lean on chromosomal/epigenetic alterations rather than on a recurrent driver mutation. Notably, chromatin remodeling and DNA repair genes, albeit different from those specific for PanNETs, have been implicated also in lung NETs by recent genomic studies (Fernandez-Cuesta et al. 2014, Simbolo et al. 2017).
As for chromosomal alterations, several studies have been published and showed a high degree of aberrations in PanNETs; these included frequent loss of chromosome 1q, 3p (where the VHL gene is located) and 11q (where MEN1 and also ATM, another pivotal gene activated by DNA double-strand breaks and involved in both damage checkpoint and double-strand repair, reside). Other losses were frequent but showed less concordance among different studies, including chromosome 6q, 10q (where PTEN resides) and 11p. Tumours also showed recurrent gains at chromosome 7q and 9q, while variable gains were located at chromosome 7p, 9p, 17q and 20q, as reviewed in Capurso et al. (2012). Meanwhile, DNA methylation studies have shown frequent hyper-methylation in a large fraction of PanNETs and identified several target genes/promoters including RASSF1A, CDKN2A and VHL. Hypo-methylation was also reported for ALU and LINE1, the latter being correlated with poor prognosis and chromosomal instability of ATRX/DAXX-negative tumours (Stefanoli et al. 2014, Di Domenico et al. 2017, Marinoni et al. 2017). This combination of diverse genetic and epigenetic alterations may be puzzling to solve but could result in more homogeneous downstream effects; therefore, researches also tackled the gene expression profiling of PanNETs. Both miRNA and mRNA profiles of primary tumours were analysed by separate studies. The study of miRNA allowed to identify a set of miRNAs that distinguish neuroendocrine from acinar tumours, also revealing the association between overexpression of miR-21 (that sustains PI3K/mTOR pathways through inhibition of PTEN) and PanNETs with high proliferation index and liver metastasis (Roldo et al. 2006, Karpathakis et al. 2013).
The PI3K/mTOR pathway was again implicated when analysing the expression profiles of PanNETs, with a particularly evident association between low PTEN and TSC2 expression levels and development of metastasis, tumour progression and poor overall survival (Missiaglia et al. 2010). This study also showed the in vitro efficacy of mTOR inhibition using PanNET cell lines that displayed reduced levels of PTEN and TSC2. More recently, an integrated study was performed in the attempt to subgroup PanNETs by simultaneous comparison of miRNA, mRNA and mutation profiles and by cross-comparing human tumours with those arising in the Rip1Tag2 mouse model of pancreas neuroendocrine tumours (Sadanandam et al. 2015). Three subtypes of tumour were identified by miRNA/mRNA profiles of both human and murine specimens: insulinoma-like tumours (IT), intermediate MEN1-like tumours and metastasis-like primary tumours (MLP). However, while IT and MLP tumours from human and mouse clustered together, MEN1-like tumours formed a compact subgroup that clustered far from the mouse model and were enriched in mutations of chromatin remodeling genes (MEN1, DAXX, ATRX). By contrast, IT tumours were enriched in mutations affecting the PI3K/mTOR pathway (TSC2, PTEN), while in the MLP subtype, both chromatin remodeling and PI3K/mTOR were affected (Sadanandam et al. 2015). Each of the previous studies, even those with the larger throughput and number of cases, could provide only a partial view of the complex molecular dysregulation that drivers PanNET oncogenesis. Furthermore, despite the apparent implication of the PI3K/mTOR pathways in at least a subset of PanNETs, a rationale for stratification of patients that could benefit from everolimus or other mTOR inhibitors could not be reached, and the hyperactivation of the mTORC2 complex in reaction to mTORC1 inhibition (Kim et al. 2017) in PI3K/mTOR dysregulated PanNETs has not been fully dissected yet. The ICGC consortium was specifically born to coordinate large worldwide studies that could collect and process high quality homogeneous data from whole genome, exome, transcriptome, microarray genomic hybridisation and ancillary techniques, on a large number of carefully verified tumour samples for each cancer type to be profiled (International Cancer Genome Consortium2010). The data produced from each project not only can confirm or confute previous data or produce new insights, but also can offer a scaffold to recompose sparse reports as in the case of rare neoplasms.
The genomic landscape of PanNETs according to the ICGC data
The ICGC whole-genome study was aimed at studying well-differentiated PanNETs, mostly of G1–G2 grade, and used 160 cases (98 in the discovery cohort and 62 in the validation cohort), excluding poorly differentiated pancreatic neuroendocrine carcinomas, mixed adenoneuroendocrine carcinomas and familial cases. Mutational analysis and comparison with previous data on pancreatic ductal adenocarcinoma (Waddell et al. 2015) confirmed the low mutational load of PanNETs. The first novel finding of the ICGC study is the definition of mutational signatures that characterise these tumours, which includes 4 processes already described in other tumours (APOBEC, age, BRCA and the unknown aetiology ‘Signature 5’) (Alexandrov et al. 2013) and a previously unreported base excision-repair (BER) deficiency signature due to the biallelic inactivation of the MUTYH gene due to pathogenic germline mutations and somatic loss of heterozygosity; this signature has been shown therein to be the same of MUTYH-associated polyposis (MAP) of the colon (Scarpa et al. 2017), and this finding has been confirmed by a recent publication that also reported the same signature in a subset of adrenocortical carcinomas (Pilati et al. 2017). The second novel finding relies in the higher than expected prevalence of germline mutations, which were found in 17% of patients lacking a family or personal history of cancer and affected the known MEN1, VHL and CDKN1B genes, and the previously unreported DNA damage repair genes MUTYH, CHEK2 and BRCA2. The third novel finding is the identification of novel mutational mechanisms including a pattern of chromosomal rearrangements compatible with chromothripsis in 9% of cases and EWSR1 gene fusions (known as a driver alteration in Ewing’s sarcoma) in 3% of tumours. The fourth relevant observation regards the analysis of somatically altered driver genes that confirmed a heavy involvement of the chromatin remodeling and PI3K/mTOR pathway in PanNETs development, with MEN1 (36 cases) as the top driver gene, DAXX (22 cases) and ATRX (10 mutated, one rearranged case) mutually exclusive alterations as the second main event and linked to ALT, followed by mutations in members of the PI3K/mTOR pathway as PTEN (7 cases), TSC1/2 (3 cases) and the previously undetected DEPDC5 (2 cases), which is part of the GATOR1 complex inhibiting mTORC1 activator Rag (Bar-Peled et al. 2013). The inactivation of SETD2, a histone modifier tumour suppressor gene (Li et al. 2016) in 6% of cases (mutation in 5 cases and rearrangement in one case) is another previously unreported recurrent alteration in PanNETs. Other chromatin remodeling genes previously reported as inactivated in lung NETs (Fernandez-Cuesta et al. 2014, Simbolo et al. 2017) were inactivated by structural rearrangements, namely ARID2 (5 cases), SMARCA4 (3 cases) and KMT2C/MLL3 (3 cases). The PI3K/mTOR pathway was involved by three novel findings: the rearrangement leading to fusion transcripts of the EWSR1 gene (3 cases), a possible marker for PI3K/mTOR targeted therapy (Giorgi et al. 2015) and the recurrent amplification of PI3K’s activator PSPN and mTOR’s regulator ULK1. The fifth relevant finding relates to RNAseq clustering of 30 cases, which confirmed the 3 subgroups previously described (Sadanandam et al. 2015) showing a prominent involvement of hypoxia-related genes in the MLP subgroup.
Four core pathways (Fig. 1) of PanNETs dominated by the presence of altered MEN1, outlined by previous research, emerge from this report with a better definition of genes involved as players, including DNA damage repair, chromatin remodeling, telomere alteration and the PI3K/mTOR signalling pathway. DNA damage repair was altered by germline mutations more often than expected, opening a question on the actual heritability level in these tumours; moreover, frequent alterations of the HR complex open the door to evaluate the applicability of therapeutic strategies (platinum or PARP-inhibitors) already approved for other HR-deficient cancers. Chromatin remodeling alterations involved not only MEN1, that was however the dominant driver in most cases, but also other complexes such as the SWI/SNF and the histone methylase complex whose alterations PanNETs share with lung NETs. This may lead to postulate a common ground for chromatin reprogramming in NETs of different districts, also opening a chapter of PanNETs research that deserves further studies to bridge genetic and epigenetic alterations. Telomere alteration by DAXX/ATRX inactivation was confirmed, while the effect of MEN1 alteration showed a more variable effect, compatible with the many functions of menin itself. DAXX/ATRX mutations also confirmed their value as predictor of poor prognosis: subjects bearing tumours with inactive DAXX/ATRX had a lower disease-specific survival, as also outlined by another recent study on 321 PanNET patients that reported an association between ALT-positive, DAXX/ATRX-negative PanNETs and shorter disease-free survival (Singhi et al. 2017). The landscape of PI3K/mTOR pathway alterations was enriched by new potential players, like DEPDC5 and EWSR1 fusions, and correlated with both the presence of DAXX/ATRX mutation and poor prognosis, a further aid for patients stratification that should be validated with larger cohorts to define the best combination of alterations with the highest independent predictive value. Moreover, the novel information on the dysregulation of the PI3K/mTOR pathway may be used as a source for biomarkers that should be tested in selected patients (e.g. those with EWSR1 fusion genes) as predictors of response in current and future clinical trials with drugs targeting this pathway.
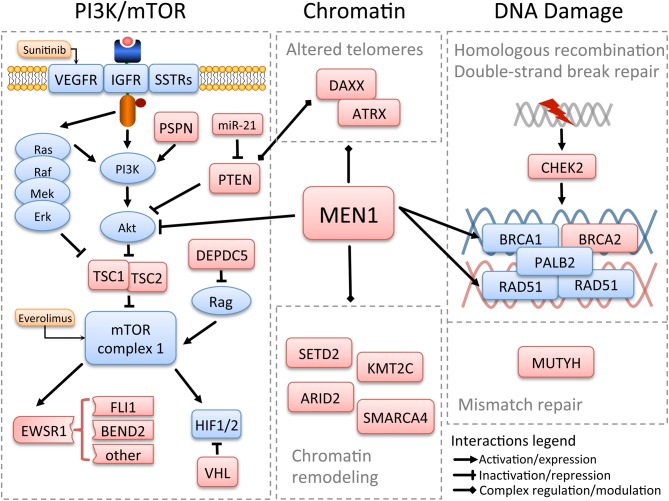
Figure 1.
Outline of main altered pathways in pancreatic neuroendocrine tumours. The scheme merges data from the literature and published data of the International Cancer Genome Consortium (ICGC). Pathway members whose genetic alteration has been proven are shaded in red, approved targeted drugs are shaded in orange. MEN1 interacts and modulates all core pathways acting as a hub gene. DAXX/ATRX also cooperate with the other genes of the chromatin remodeling complexes. DAXX modulates PTEN distribution between the nucleus and the cytoplasm (Song et al. 2008), and the modulation of DAXX by PTEN has been reported soon after publication of the ICGC data (Benitez et al. 2017).
Conclusion
The ICGC data on PanNETs allowed to draw the first snapshot of the heterogeneous combination of genetic alterations that drive this type of tumours. Despite this variability, some key pathways are consistently targeted by the tumorigenic process, resulting in a frequent loss of MEN1 function, activation of the PI3K/mTOR pathway, chromatin remodeling and telomeres alteration. While the next step for translational research will be the integration of genomic, transcriptomic and epigenomic data to identify key bottlenecks of this network and to explain how different alterations lead to a small number of subgroups according to expression profiles, these data may propel clinical research aimed at identifying and validating efficient markers for patients stratifications and to guide the choice of targeted therapeutic options, as recently outlined (Capdevila et al. 2017).
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Funding
This work was supported in part by Associazione Italiana per la Ricerca sul Cancro (grant n.12182); European Commission Seventh Framework Programme FP7 Health (Cam-Pac, grant agreement 602783).
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al 2013. Signatures of mutational processes in human cancer. Nature 500 415–421. ( 10.1038/nature12477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. 2013. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340 1100–1106. ( 10.1126/science.1232044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez JA, Ma J, D’Antonio M, Boyer A, Camargo MF, Zanca C, Kelly S, Khodadadi-Jamayran A, Jameson NM, Andersen M, et al 2017. PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nature Communications 8 15223 ( 10.1038/ncomms15223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Casanovas O, Salazar R, Castellano D, Segura A, Fuster P, Aller J, Garcia-Carbonero R, Jimenez-Fonseca P, Grande E, et al 2017. Translational research in neuroendocrine tumors: pitfalls and opportunities. Oncogene 36 1899–1907. ( 10.1038/onc.2016.316) [DOI] [PubMed] [Google Scholar]
- Capurso G, Festa S, Valente R, Piciucchi M, Panzuto F, Jensen RT, Delle Fave G. 2012. Molecular pathology and genetics of pancreatic endocrine tumours. Journal of Molecular Endocrinology 49 R37–R50. ( 10.1530/JME-12-0069) [DOI] [PubMed] [Google Scholar]
- Corbo V, Dalai I, Scardoni M, Barbi S, Beghelli S, Bersani S, Albarello L, Doglioni C, Schott C, Capelli P, et al 2010. MEN1 in pancreatic endocrine tumors: analysis of gene and protein status in 169 sporadic neoplasms reveals alterations in the vast majority of cases. Endocrine-Related Cancer 17 771–783. ( 10.1677/ERC-10-0028) [DOI] [PubMed] [Google Scholar]
- Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, Shih T, Yao JC. 2017. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncology 3 1335–1342. ( 10.1001/jamaoncol.2017.0589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde RF, Heaphy CM, Maitra A, Meeker AK, Edil BH, Wolfgang CL, Ellison TA, Schulick RD, Molenaar IQ, Valk GD, et al 2012. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Modern Pathology 25 1033–1039. ( 10.1038/modpathol.2012.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domenico A, Wiedmer T, Marinoni I, Perren A. 2017. Genetic and epigenetic drivers of neuroendocrine tumours (NET). Endocrine-Related Cancer 24 R315–R334. ( 10.1530/ERC-17-0012) [DOI] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Peifer M, Lu X, Sun R, Ozretic L, Seidal D, Zander T, Leenders F, George J, Muller C, et al 2014. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nature Communications 5 3518 ( 10.1038/ncomms4518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajate P, Martinez-Saez O, Alonso-Gordoa T, Grande E. 2017. Emerging use of everolimus in the treatment of neuroendocrine tumors. Cancer Management and Research 9 215–224. ( 10.2147/CMAR.S113382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Boro A, Rechfeld F, Lopez-Garcia LA, Gierisch ME, Schafer BW, Niggli FK. 2015. PI3K/AKT signaling modulates transcriptional expression of EWS/FLI1 through specificity protein 1. Oncotarget 6 28895–28910. ( 10.18632/oncotarget.5000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al 2011. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333 425 ( 10.1126/science.1207313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Cancer Genome Consortium, Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, Bhan MK, Calvo F, Eerola I, et al 2010. International network of cancer genome projects. Nature 464 993–998. ( 10.1038/nature08987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331 1199–1203. ( 10.1126/science.1200609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpathakis A, Dibra H, Thirlwell C. 2013. Neuroendocrine tumours: cracking the epigenetic code. Endocrine-Related Cancer 20 R65–R82. ( 10.1530/ERC-12-0338) [DOI] [PubMed] [Google Scholar]
- Kim LC, Cook RS, Chen J. 2017. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 36 2191–2201. ( 10.1038/onc.2016.363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. PNAS 107 14075–14080. ( 10.1073/pnas.1008850107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. 2016. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget 7 50719–50734. ( 10.18632/oncotarget.9368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R, Osamura RY, Klöppel G, Rosai J. 2017. WHO Classification of Tumours of Endocrine Organs. Lyon, France: IARC Press. [Google Scholar]
- Marinoni I, Kurrer AS, Vassella E, Dettmer M, Rudolph T, Banz V, Hunger F, Pasquinelli S, Speel EJ, Perren A. 2014. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology 146 453.e455–460.e455. ( 10.1053/j.gastro.2013.10.020) [DOI] [PubMed] [Google Scholar]
- Marinoni I, Wiederkeher A, Wiedmer T, Pantasis S, Di Domenico A, Frank R, Vassella E, Schmitt A, Perren A. 2017. Hypo-methylation mediates chromosomal instability in pancreatic NET. Endocrine-Related Cancer 24 137–146. ( 10.1530/ERC-16-0554) [DOI] [PubMed] [Google Scholar]
- Missiaglia E, Dalai I, Barbi S, Beghelli S, Falconi M, della Peruta M, Piemonti L, Capurso G, Di Florio A, delle Fave G, et al 2010. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. Journal of Clinical Oncology 28 245–255. ( 10.1200/JCO.2008.21.5988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati C, Shinde J, Alexandrov LB, Assie G, Andre T, Helias-Rodzewicz Z, Ducoudray R, Le Corre D, Zucman-Rossi J, Emile JF, et al 2017. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. Journal of Pathology 242 10–15. ( 10.1002/path.4880) [DOI] [PubMed] [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, et al 2006. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. Journal of Clinical Oncology 24 4677–4684. ( 10.1200/JCO.2005.05.5194) [DOI] [PubMed] [Google Scholar]
- Sadanandam A, Wullschleger S, Lyssiotis CA, Grotzinger C, Barbi S, Bersani S, Korner J, Wafy I, Mafficini A, Lawlor RT, et al 2015. A cross-species analysis in pancreatic neuroendocrine tumors reveals molecular subtypes with distinctive clinical, metastatic, developmental, and metabolic characteristics. Cancer Discovery 5 1296–1313. ( 10.1158/2159-8290.CD-15-0068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa A, Corbo V, Barbi S, Cataldo I, Fassan M. 2015. Molecular biology of neuroendocrine tumors. In Neuroendocrine Tumours: Diagnosis and Management. pp 35–49. Eds Yalcin S. & Oberg K. Berlin, Heidelberg, Germany: Springer-Verlag; ( 10.1007/978-3-662-45215-8_1) [DOI] [Google Scholar]
- Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini A, et al 2017. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543 65–71. ( 10.1038/nature21063) [DOI] [PubMed] [Google Scholar]
- Scherubl H, Streller B, Stabenow R, Herbst H, Hopfner M, Schwertner C, Steinberg J, Eick J, Ring W, Tiwari K, et al 2013. Clinically detected gastroenteropancreatic neuroendocrine tumors are on the rise: epidemiological changes in Germany. World Journal of Gastroenterology 19 9012–9019. ( 10.3748/wjg.v19.i47.9012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbolo M, Mafficini A, Sikora KO, Fassan M, Barbi S, Corbo V, Mastracci L, Rusev B, Grillo F, Vicentini C, et al 2017. Lung neuroendocrine tumours: deep sequencing of the four World Health Organization histotypes reveals chromatin-remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. Journal of Pathology 241 488–500. ( 10.1002/path.4853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhi AD, Liu TC, Roncaioli JL, Cao D, Zeh HJ, Zureikat AH, Tsung A, Marsh JW, Lee KK, Hogg ME, et al 2017. Alternative lengthening of telomeres and loss of DAXX/ATRX expression predicts metastatic disease and poor survival in patients with pancreatic neuroendocrine tumors. Clinical Cancer Research 23 600–609. ( 10.1158/1078-0432.CCR-16-1113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. 2008. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455 813–817. ( 10.1038/nature07290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanoli M, La Rosa S, Sahnane N, Romualdi C, Pastorino R, Marando A, Capella C, Sessa F, Furlan D. 2014. Prognostic relevance of aberrant DNA methylation in g1 and g2 pancreatic neuroendocrine tumors. Neuroendocrinology 100 26–34. ( 10.1159/000365449) [DOI] [PubMed] [Google Scholar]
- Swarts DR, Scarpa A, Corbo V, Van Criekinge W, van Engeland M, Gatti G, Henfling ME, Papotti M, Perren A, Ramaekers FC, et al 2014. MEN1 gene mutation and reduced expression are associated with poor prognosis in pulmonary carcinoids. Journal of Clinical Endocrinology and Metabolism 99 E374–E378. ( 10.1210/jc.2013-2782) [DOI] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al 2015. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518 495–501. ( 10.1038/nature14169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, Abdalla EK, Fleming JB, Vauthey JN, Rashid A, et al 2008. One hundred years after ‘carcinoid’: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. Journal of Clinical Oncology 26 3063–3072. ( 10.1200/JCO.2007.15.4377) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a