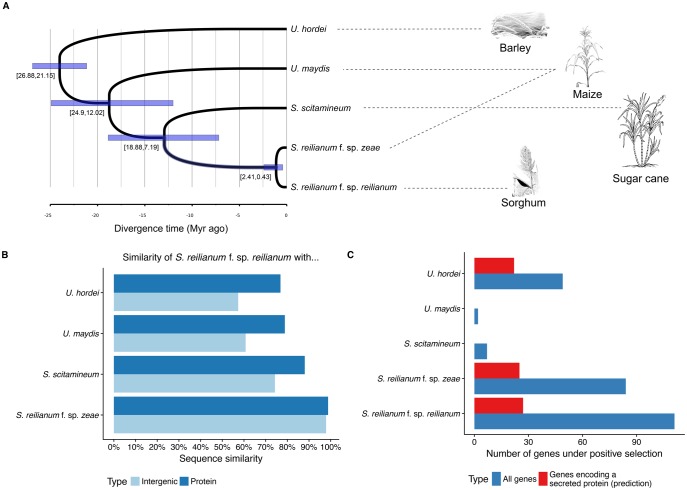
Fig. 1.
—Phylogeny, divergence estimates, and number of genes under positive selection in five related smut fungal species parasitizing different host plants. (A) Chronogram of the five fungal pathogens as estimated under a relaxed molecular clock. Boxes represent 95% posterior intervals, with corresponding values indicated below. (B) Pairwise sequence differences, for both the noncoding genome and the proteome (nonsynonymous differences). (C) Number of positively selected genes on each terminal branch (total number of genes and genes predicted to encode a secreted protein).