Fig. 3. Runx2 mRNA accumulates during mitosis and segregates to progeny cells following cell division.
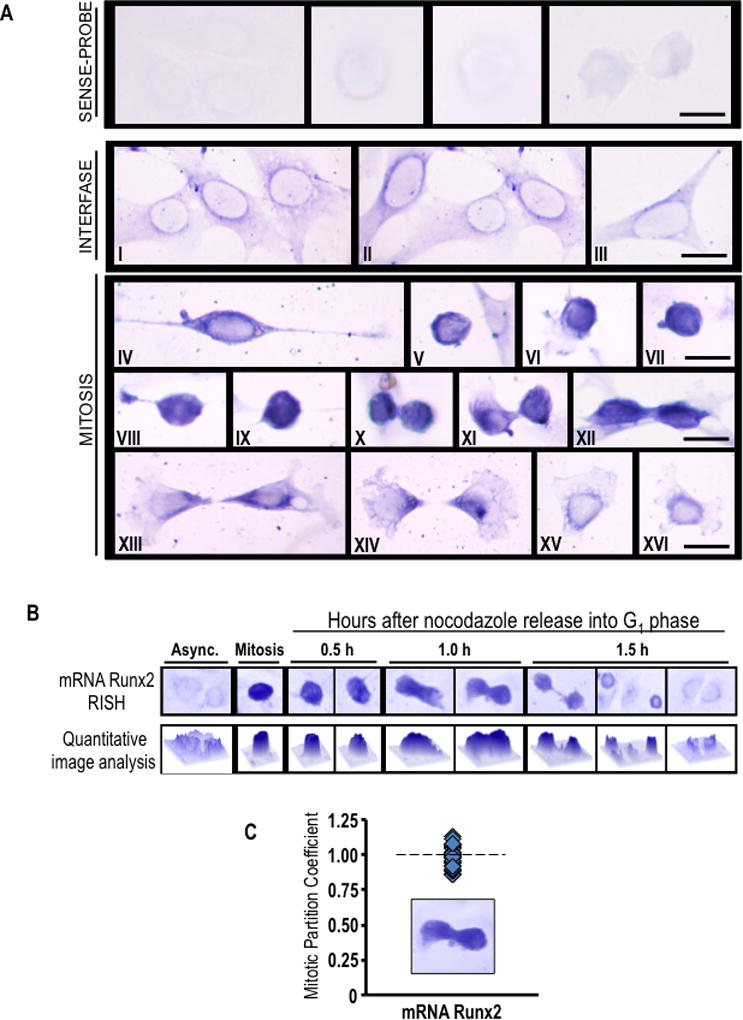
(A) Asynchronously growing MC3T3-E1 cells were fixed and subjected to in situ hybridization (RISH) analysis of Runx2 mRNA using DIG-labeled antisense probe. Sense probe was used as a control. Cytoplasmic/perinuclear blue staining denotes presence of Runx2 mRNA in pre-osteoblast cells during interphase (I-III). As interphase cells progressing into mitosis, Runx2 mRNA is concentrated as strong blue-black staining that is observed throughout the cell cortex (IV-IX). At cytokinesis, most of Runx2 mRNA persists in a cortical distribution (X-XII). After cell division, Runx2 mRNA appears to be segregated and evenly distributed between daughter cells (XIII-XIV) showing a polarized distribution at the region of the cleavage furrow. When the remaining cytoplasm of the midbody is retracted into nascent cells, Runx2 mRNA relocalizes around the nucleus (XV-XVI). Interphase and mitotic cells were identified by cell morphology. Scale bars: 20 µm, 100X oil objective, numerical aperture = 1.25. (B and C) RISH studies were also performed during mitotic block-release experiments at specific time points (0.5, 1.0 and 1.5 h) after nocodazole release. Runx2 RISH signal intensity was examined in situ by microscopy and quantitative image analysis (B) of progeny cells at the last step of cytokinesis (n=10) (C). We defined the partition coefficient, which reflects the ratio of integrated signal intensities between progeny cells. Cells progressing into G1 exhibit symmetrical partitioning of Runx2 mRNA between daughter cells.
