Figure 2.
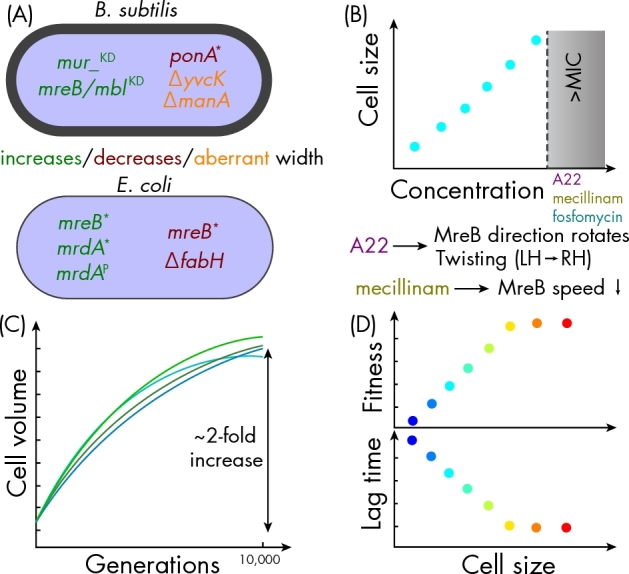
Molecular and evolutionary changes to cell width. (A) Genes with established connections to cell-width determination in B. subtilis (top) and E. coli (bottom). *: mutation in coding region; KD: CRISPRi knockdown; P: promoter mutation. Color indicates whether perturbation leads to increased, decreased, or aberrant cell width. (B) Sublethal treatments with A22 (targets MreB), mecillinam (PBP2), and fosfomycin (MurA) increase cell width. For A22, increases in cell width are correlated with rotation of the direction of MreB movement and a continuous transition from left-handed to right-handed twisting, while mecillinam causes MreB speed to decrease (Tropini et al.2014). (C) Cell volume was observed to increase concurrently with cell fitness in a long-term evolution experiment (Lenski and Travisano 1994). Each line in the schematic represents an individual evolved population. (D) The increased cell width of mreBA53X mutants is correlated with increased competitive fitness and decreased lag time (Monds et al.2014).
