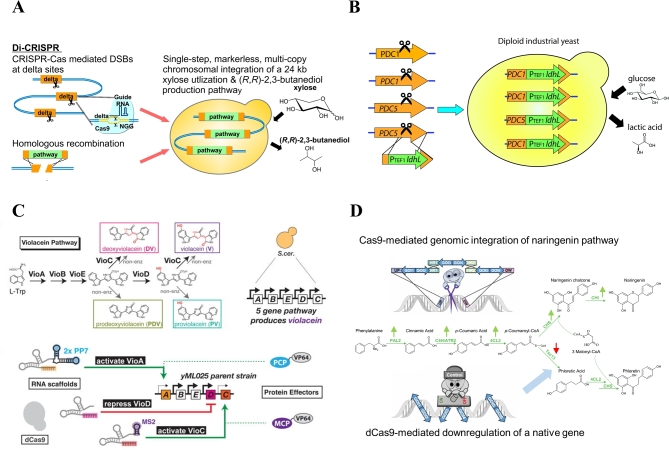
Figure 4.
Application of CRISPR/Cas9 systems for engineering of yeast cell factories. (A) Production of (R,R)-2,3-butanediol from xylose. Multicopy one-step integration of the xylose utilization and (R,R)-2,3-butanediol pathways into Ty-element delta sites in the genome (The figure is reprinted with permission from Elsevier: Shi et al. A highly efficient single-step, markerless strategy for multicopy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab Eng 2016;33:19–27.). (B) Production of lactic acid from glucose in an industrial yeast strain, one-step disruption of two genes in diploid strain and simultaneous integration of lactate dehydrogenase genes from L. plantarum (ldhL) (Stovicek, Borodina and Forster 2015). (C) Production of deoxyviolacein, violacein, prodeoxyviolacein and proviolacein from glucose. Transcriptional regulation (activation/repression) of different genes in violacein pathway leads to production of different violacein derivatives (The figure is reprinted with permission from Elsevier: Zalatan et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 2015;160:339–50.): VP64-activator domain, PP7/MS2 – RNA hairpin structures, PCP/MCP—RNA binding proteins. (D) Production of naringenin from glucose. Cas9-mediated one-step integration of the naringenin pathway into an intergenic locus. Downregulation of TSC13 mediated by catalytically inactive (‘dead’) dCas9 (CRISPRi) to avoid the formation of by-products (The figure adapted from Vanegas, Lehka and Mortensen 2017).