Abstract
The main focus in development of yeast cell factories has generally been on establishing optimal activity of heterologous pathways and further metabolic engineering of the host strain to maximize product yield and titer. Adequate stress tolerance of the host strain has turned out to be another major challenge for obtaining economically viable performance in industrial production. Although general robustness is a universal requirement for industrial microorganisms, production of novel compounds using artificial metabolic pathways presents additional challenges. Many of the bio-based compounds desirable for production by cell factories are highly toxic to the host cells in the titers required for economic viability. Artificial metabolic pathways also turn out to be much more sensitive to stress factors than endogenous pathways, likely because regulation of the latter has been optimized in evolution in myriads of environmental conditions. We discuss different environmental and metabolic stress factors with high relevance for industrial utilization of yeast cell factories and the experimental approaches used to engineer higher stress tolerance. Improving stress tolerance in a predictable manner in yeast cell factories should facilitate their widespread utilization in the bio-based economy and extend the range of products successfully produced in large scale in a sustainable and economically profitable way.
Keywords: stress tolerance, metabolic engineering, cell factory, Saccharomyces cerevisiae
Engineering stress tolerance into yeast cell factories is key to their successful application.
INTRODUCTION
There has been a surge of interest in the commercial production of bio-based chemicals with microorganisms used as cell factories (Becker and Wittmann 2012; Chung et al.2015; Tsuge et al.2016). The yeast Saccharomyces cerevisiae has been a favorite organism in this respect because of its long-standing use in classical industrial applications, such as beer and wine production, its extensive toolbox for genetic modification and the vast knowledge on its physiology, molecular biology and genetics (Kampranis and Makris 2012).
Although industrial yeast strains have great robustness, they often lack tolerance to specific stress factors when used as cell factories. A first near-universal stress factor is the toxicity of the end product, which has to be accumulated with high yield and maximal titer in order to ensure economic viability of the industrial process. Favored chemicals for cell factory production like organic acids exert strong inhibition on the metabolism of microorganisms, including yeast (Narendranath, Thomas and Ingledew 2001). Even the accumulation of very high levels of ethanol in biofuel production is toxic to the yeast (Stanley et al. 2010a; Pais et al.2013). A second source of stress factors is the composition of the substrate and its pretreatment process. Pure sugar streams can be used as feedstock for cell factory production of bio-based chemicals, but because of the low value of bulk chemicals and biofuels, the use of cheaper substrates is preferred. The latter are usually much more heterogeneous and often contain high levels of inhibitors, either present in the substrate itself or generated during the pretreatment process (Palmqvist and Hahn-Hägerdal, 2000). This necessitates the engineering of much higher tolerance against these inhibitors than is generally present in natural or industrial strains of species used as cell factories, including yeast. A third major stress factor is high temperature. Enzymatically catalyzed reactions and thus also microbial production processes for bio-based compounds proceed faster at higher temperature, which is favored because it enhances the productivity of the commercial plant and thus reduces capital expenses (capex). In addition, microbial fermentation processes are exergonic. They produce heat, and large-scale fermentors thus have to be properly cooled. In combination with changing environmental temperatures, this can cause temperature gradients and fluctuations in the fermentors that can compromise fermentation rate and productivity (Abdel-Banat et al.2010). Additional stress factors include high osmolarity. Because of the need to achieve high product titers, very high gravity fermentations are required, increasing osmotic pressure during fermentation. Salt tolerance can be important because of high salt levels introduced during the pretreatment, the use of new feedstocks such as seaweed, the cleaning and water recirculation practices (Maiorella, Blanch and Wilke 1984; Silverstein et al.2007; Wi et al.2009; Chavez-Rodriguez et al.2013; Wei, Quarterman and Jin 2013). These stress factors not only require a high level of general robustness of the cell factory microorganism but also much higher tolerance to specific stress factors than usually present even in the most robust industrial strains. The stress factors very often also reinforce each other making it even more difficult to reach the required level of fermentation performance under real industrial conditions.
A rather unexpected outcome of the development of microbial cell factories is that the new artificially engineered metabolic pathways tend to be much more sensitive to stressful conditions than the intrinsic metabolic pathways of the organism. This is very clear for instance in the co-fermentation of glucose and xylose in second-generation bioethanol production, in which the artificially engineered xylose fermentation turns out to be much more sensitive to inhibitors such as acetic acid compared to glucose fermentation (Bellissimi et al.2009). This is likely due to the extensive adaptation and selection that the microorganism has undergone during evolution when fermenting its natural substrates under myriads of different environmental conditions, whereas fermentation of the artificial substrate has never undergone a similar fine-tuned integration in the regulatory network governing microbial metabolism.
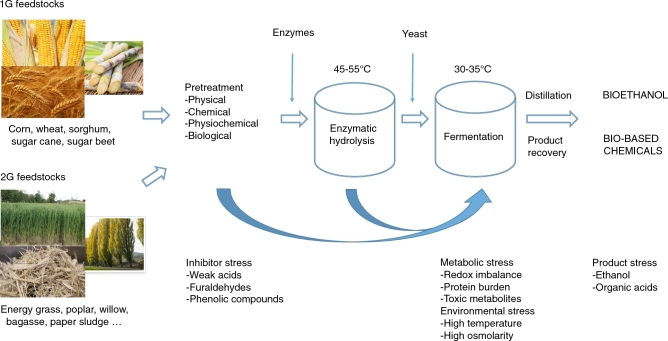
This review provides an overview of the most important stress factors for yeast cell factories in industrial production processes (Fig. 1). Many of these stress conditions are general for all fermentations. However, due to the nature of the biomass and the pretreatment process used in second-generation bioethanol production, yeast cell factories are confronted with several additional stress factors in this process.
Figure 1.
Schematic overview of the different steps in the industrial production of bioethanol and bio-based chemicals with first- and second-generation substrates using yeast cell factories and the most common stress factors associated with the different subprocesses.
To improve the yields and titers, superior alleles conferring tolerance to specific stress factors could be engineered in industrial strains using several methods, with one of the most powerful the recently developed, highly precise and scarless CRISPR-Cas9 genome editing approach.
Also, several non-conventional yeast species display specific properties that are highly desirable in the fermentation industry such as thermotolerance, weak acid tolerance and osmotolerance (Kluyveromyces marxianus, Zygosaccharomyces bailii and Debaryomyces hansenii, respectively) (Radecka et al.2015). However, despite the superior growth of these yeast species under the specific stress condition, their implementation in industrial fermentations is often troublesome due to absent or very limited fermentation capacity and a low general robustness.
RELEVANT STRESS FACTORS
Industrial production processes put a multitude of stresses on yeast cell factories. Physical and chemical extracellular stresses include temperature, osmotic pressure and the presence of lignocellulosic biomass-derived inhibitors. Furthermore, producing recombinant proteins in yeast cell factories can create a protein burden on the cell and engineering novel metabolic pathways can cause redox imbalances.
Environmental stress factors
Alcoholic fermentation is a well-established industrial process where several factors can affect yeast performance and most of them are also relevant in other bio-based fermentations. For many of the abovementioned stress factors, genes that confer resistance have already been identified. However, most of these studies were performed with lab strains and it is not clear to what extent the genes identified can be used to increase the tolerance of industrial strains.
High ethanol concentration
In order to make the distillation of ethanol economically viable, industry usually requires a minimal concentration of 4%–5% (v/v) ethanol. Lower ethanol concentrations dramatically increase the energy demand for distillation (Stampe et al.1983; Zacchi and Axelsson 1989). In the USA, corn ethanol production often reaches a final ethanol concentration of 16%–18%, while Brazilian sugar cane ethanol falls in the range of 8%–11% (Shapouri and Gallagher 2005; Basso et al.2008). The production processes are becoming more advanced resulting in higher ethanol yields and titers, and the industry is thus in need of yeast strains with higher maximal ethanol accumulation capacity.
Ethanol has many effects on cell growth and viability. It alters membrane composition and fluidity (Jimenez and Benítez 1987; Chi and Arneborg 2000; You, Rosenfield and Knipple 2003; Takagi et al.2005; Huffer et al.2011; Henderson and Block 2014), which results in deregulated flux of ions, especially protons. This reduces the proton-motive force over the membrane (Cartwright et al.1986), compromising nutrient uptake through the many proton symporters that are active in yeast. Also, divalent ions such as Mg2+ are of importance. It has been shown that magnesium is not only important as cofactor during fermentation, but also as ethanol protectant (Walker and Maynard 1997). Additionally, ethanol reduces the activity of the plasma membrane H+-ATPase (Cartwright, Veazey and Rose 1987; Aguilera et al.2006) and of glycolytic enzymes due to reduced water activity (Hallsworth, Nomura and Iwahara 1998; Ogawa et al.2000; Alexandre et al.2001). Another possible consequence of ethanol toxicity is the loss of mitochondrial DNA (Ibeas and Jimenez 1997).
Many studies on ethanol tolerance were carried out using DNA microarrays in which either the genome-wide gene expression levels of a strain in the presence or in the absence of ethanol stress (7% (Alexandre et al.2001), 5% (Hirasawa et al.2007)) or those of an ethanol tolerant and an ethanol-sensitive strain under the same conditions were compared (Watanabe et al. 2007c; Dinh et al.2009; Lewis et al.2010). Screening of the Saccharomyces cerevisiae gene deletion collection resulted in identification of 46 mutants with higher sensitivity to ethanol (van Voorst et al.2006). Swinnen et al. (2012) identified two causative genes for high ethanol tolerance (MKT1 and APJ1) in S. cerevisiae VR1–5B, a haploid segregant of the very ethanol tolerant, industrial S. cerevisiae strain VR1. As inferior parent, the lab strain BY4741, in which the MKT1 gene is defective, was used. The VR1–5B MKT1 allele rendered the strain BY4741 more ethanol tolerant, but may not be very useful for further improving ethanol tolerance in industrial strains since they do have already a wild-type MKT1 allele. This experience underscores the importance of using industrial or natural strains for identification of relevant (mutant) genes that can improve a specific trait of interest in industrial production strains.
High osmotic pressure and ion toxicity
Osmotic stress in bioethanol production is caused by high sugar concentrations and/or high salt concentrations. In first-generation bioethanol production, osmotic pressure is mostly caused by the concentrated streams of about 35% sugar that are used and are required to reach ethanol titers of 16%–18% (Bertrand et al.2016). In second-generation bioethanol production, it is difficult to reach high concentrations of fermentable sugars. They are usually limited to about 12% sugar so that an ethanol titer of only 5%–7% is reached. Hence, osmotic pressure caused by high sugar concentration is usually not a problem in second-generation bioethanol production. On the other hand, only part of the solids can be converted into fermentable sugars. Increased solids loading requires additional pumping power and proper mixing is impaired when the viscosity in the reactor is too high (Fan et al.2003). At high solids loading, the cellulolytic enzymes also start to become inhibited (Kristensen, Felby and Jorgensen 2009). Solids mainly affect the enzymatic hydrolysis because of enzyme adsorption to their surface while they have less effect on the yeast fermentation.
High sodium salt concentrations present in the second-generation bioethanol production process, due to the pretreatment method or to procedures used to detoxify the hydrolysate, could also compromise the fermentation rate and yield. Casey et al. (2013) investigated the effect of different salts on glucose-xylose co-fermentation by S. cerevisiae. Their results indicate that the different salts do not affect final yield, but instead reduce the sugar consumption rate. In this respect, the xylose consumption rate is more affected by higher salt concentrations than the glucose consumption rate. Another observation is that salt toxicity is less dependent on the type of cation present (Na+, K+ or NH4+) and more on the type of anion (SO42− is more toxic than Cl−). However, the presence of certain cations does seem to influence the growth rate of S. cerevisiae differentially. While sodium and calcium salts have a similar, strongly inhibitory effect on yeast growth, potassium and magnesium salts affect yeast growth to a lesser extent (Bautista-Gallego et al.2008).
Removing the salt is not a good option because of the costs involved (Sanchez and Cardona 2008). Furthermore, wastewater treatment and water recycling practices can also result in accumulation of (sodium) salt in pipelines and fermentors. The different salts do not only cause osmotic stress to the cell, but can also cause ion toxicity (Ranatunga et al.2000; Hohmann 2002; Millati, Niklasson and Taherzadeh 2002; Hohmann, Krantz and Nordlander 2007; Wei, Quarterman and Jin 2013).
Some of the feedstocks contain, besides potassium and sodium salts, also (heavy) metal salts. Sugarcane bagasse, for instance, usually contains Mg2+and Fe2+ salts and other trace elements of Mn2+, Cu2+ and Zn2+ compounds (Wythes, Wainwright and Blight 1978; Kaushal et al.1981). Although manganese, copper and zinc are essential for cellular function, an excessive amount is toxic for the cell. In the comprehensive work of Jin et al. (2008), the effect of different metal ions was compared for their inhibition of the growth of S. cerevisiae. Transcriptional analysis revealed that most metals reduce ribosome biosynthesis and thereby actively reduce cell growth.
High extracellular solute concentrations lead to passive water diffusion out of the cell, causing cell shrinkage and reduced intracellular water activity (Brown 1976; Cray et al.2015). Saccharomyces cerevisiae and other microorganisms can generally only grow in a very narrow range of water activity (0.990–0.998) (Brown 1976). This has enabled food producers to reduce food spoilage by actively reducing water activity (Abdullah et al.2000). Reduced water availability has also been shown to raise oxidative stress, which could lead to lipid peroxidation, protein degradation and DNA damage (Hansen, Go and Jones 2006; Garre et al.2010). Saccharomyces cerevisiae has distinct mechanisms to cope with these types of stress, but there also seems to be common mechanisms (Causton et al.2001) such as a central role for the HOG pathway, increasing intracellular osmolytes, especially glycerol and upregulation of export of toxic ions (Posas et al.2000; Tamás et al.2000; Muzzey et al.2009).
Most of the efforts to unravel the molecular-genetic basis of osmotolerance in yeast have used microarray gene expression data or physiological characterization of osmotolerant yeast species (Posas et al.2000; Schoondermark-Stolk et al.2002; Erasmus, van der Merwe and van Vuuren 2003; O’Rourke and Herskowitz, 2004). To our knowledge, none of the genes linked to osmotolerance in fundamental research have been used to engineer higher osmotolerance in industrial strains.
High temperature
Thermotolerance is another highly desirable trait in the yeast fermentation industry. This is not limited to tropical regions where temperatures can easily reach more than 40°C in the summer. Appropriate cooling of the fermentors is required because of the exergonic fermentation process (Wheals et al.1999). Higher tolerance to temperature overshoots and a higher mean fermentation temperature would provide more consistent fermentation performance, reduced contamination, lower ethanol distillation costs, a higher fermentation rate and thus reduced fermentor sizes and lower capital costs, and reduced cooling/heating expenses (Hamelinck, Hooijdonk and Faaij 2005; Stephen, Mabee and Saddler 2012). High thermotolerance is even more important in advanced processes of second-generation bioethanol production. As opposed to the regular process of separate hydrolysis and fermentation, in which enzymatic hydrolysis and yeast fermentation are carried out sequentially, in simultaneous saccharification and fermentation (SSF), enzymatic hydrolysis and yeast fermentation are carried out simultaneously. The latter process minimizes feedback inhibition by monosaccharides on enzymatic hydrolysis of the lignocellulosic polymers and also minimizes bacterial contamination because of the sustained very low free sugar levels. However, the optimum fermentation temperature of most S. cerevisiae strains is 30°C–35°C while the optimum temperature of the (ligno-)cellulolytic enzymes is 45°C–55°C (Choudhary, Singh and Nain 2016). Hence, higher thermotolerance of the yeast allowing fermentation temperatures of 40°C or higher would be highly beneficial for the SSF process. The same is true for consolidated bioprocessing, a further advanced process in which (ligno-)cellulolytic enzymes are produced by the host yeast. As a result, the amount of the enzymes needed for hydrolysis of the sugar polymers, which still amounts to 20%–30% of the cost of the ethanol, can be reduced significantly (Kawaguchi et al.2016). Obviously, also in this case a higher hydrolysis/fermentation temperature will greatly benefit the efficiency of the process.
In response to a temperature increase, yeast upregulates the transcription of heat-shock proteins (HSPs) and the accumulation of trehalose (Neves and François 1992; Ribeiro, Silva and Panek 1994; Lindquist and Kim 1996; Feder and Hofmann 1999). Whereas Hsp104 assists in correct protein folding, Hsp12 increases the integrity of the plasma membrane (Sanchez et al.1992; Sales et al.2000; Watanabe et al. 2007c; Welker et al.2010). The protective function of trehalose is most likely due to the fact that membrane and protein structures are stabilized (Wiemken 1990; Singer and Lindquist 1998; Ogawa et al.2000; Kaino and Takagi 2008; Verghese et al.2012). Increased temperatures may also lead to an increase in ROS formation in the form of hydrogen peroxide and superoxide, as well as free oxygen radicals (Moraitis and Curran 2004; Morano, Grant and Moye-Rowley 2012).
Several genes have been implicated in thermotolerance in S. cerevisiae. Overexpression of RSP5, a ubiquitin ligase (Shahsavarani et al.2012), increases thermotolerance. Enzymes involved in membrane synthesis and composition like ERG3 (Caspeta et al.2014), a C-5 sterol desaturase, chaperones such as HSP104 and HSP12 (Sanchez et al.1992), genes involved in trehalose metabolism (TPS1, TPS2 and NTH1 (De Virgilio et al.1994)) and genes of RNA processing, such as PRP42 and SMD2 (Yang et al.2013), have been linked to high thermotolerance. Nevertheless, successful industrial application of these genes in improving thermotolerance of commercial yeast strains has not been reported yet. One possible reason is that genetic modification of these genes improves thermotolerance at the expense of other properties that are important in industrial practice, either in the fermentation itself or in propagation, drying or storage of the bulk yeast.
Weak acids
The most abundant and inhibitory weak acid present in lignocellulose biomass is acetic acid. Its level varies considerably with the composition of the biomass. Second-generation feedstocks for bioethanol production have a very variable composition in terms of cellulose, hemicellulose and lignin content. Hemicellulose is a heteromeric polysaccharide consisting of substituted glucans, mannans and xylans (McNeil et al.1984; Scheller and Ulvskov 2010; Limayem and Ricke 2012). Also, the type of pretreatment process has a considerable effect on the level of acetic acid (Kristensen, Felby and Jorgensen 2009; Du et al.2010; Zha, Muilwijk and Coulier 2012).
Many pretreatment methods release acetyl groups from the hemicellulose fraction. The resulting acetic acid in unbuffered hydrolysates easily causes sluggish fermentations or may inhibit fermentation completely (Palmqvist et al.1999; Palmqvist and Hahn-Hägerdal 2000; Narendranath, Thomas and Ingledew 2001; Graves et al.2006; Bellissimi et al.2009). This is mostly due to the intracellular accumulation of protons and dramatic drop in pH caused by diffusion of the acetic acid through the plasma membrane and its dissociation at the intracellular pH which is far above its pKa of 4.76 (Guldfeldt and Arneborg 1997). Similar toxicity problems are caused by other organic acids, such as levulinic acid and formic acid, which are conversion products of hexose and pentose sugars, respectively (Horvat et al.1985; Rasmussen, Sorensen and Meyer 2014).
Acetic acid can also enter the cell through the Fps1 aquaglyceroporin channel by means of facilitated diffusion. Since Fps1 can only transport the undissociated form, and since the acetate ion can also not diffuse through the phospholipid bilayer of the plasma membrane, acetic acid toxicity is greatly reduced at an extracellular pH above its pKa (4.76) (Mollapour and Piper 2007; Ullah et al.2012). After dissociation of acetic acid in the cytoplasm, the resulting accumulation of protons and intracellular acidification strongly stimulate the activity of the proton ATPase, which consumes large amounts of ATP to expel the protons again from the cells. This results in a decrease in ATP availability, which lowers all biosynthetic processes. This is reinforced further by the decrease in proton-motive force, due to the lower intracellular pH and membrane potential, which compromises nutrient uptake and thus reduces overall metabolism including the expression of new nutrient transporters (Eraso and Gancedo 1987; Imai and Ohno 1995; Lohmeier-Vogel, Sopher and Lee 1998; Martinez-Munoz and Kane 2008; Ullah et al.2012; Ding et al.2013). The low cytosolic pH is also inhibitory to the activity of glycolytic enzymes, which weakens ATP production and thus further compromises the overall energy status of the cell (Pampulha and Loureiro-Dias 1990; Veine, Arscott and Willians 1998).
The ABC-transporter Pdr12 plays a role in acetic and levulinic acid tolerance. Its overexpression makes cells more resistant to levulinic acid, while deletion improves acetic acid tolerance. The mechanism behind this seemingly contradictory action has not yet been elucidated (Kawahata et al.2006; Ullah et al.2012; Nygård et al.2014). Also, the genes of the RIM101 pathway, a zinc-finger transcription factor, play a role in acetic acid tolerance. Targets of Rim101 include genes encoding iron transporters and proteins involved in cell wall maintenance and organization. The mechanism behind its protective function is not clear yet (Fernandes et al.2005; Mira et al.2009).
Genome-wide screens have revealed many genetic elements important for high acetic acid tolerance (Mira et al. 2010; Mira, Teixeira and Sa-Correia 2010). A major role in this respect is played by the Haa1 transcription factor that mediates expression of multiple genes required for high acetic acid tolerance (Fernandes et al.2005; Mira, Becker and Sa-Correia 2010). Several genes involved in acetic acid tolerance have been also identified by polygenic analysis, being HAA1, VMA7, GLO1, DOT5 and CUP2 (Meijnen et al.2016). In previous studies, the transcription factor HAA1 was already linked to weak acid tolerance (Fernandes et al.2005; Mira, Becker and Sa-Correia 2010), as was VMA7 (Kawahata et al.2006). Meijnen et al. (2016) identified a unique point mutation in the HAA1 gene product (G1517A) that is sufficient by itself to increase acetic acid tolerance in the xylose fermenting industrial S. cerevisiae strain GSE16-T18. The point mutation considerably reduced the lag phase of the fermentation in the presence of acetic acid. CUP2 is a paralog of HAA1 and was shown to be involved in copper tolerance (Buchman et al.1989; Welch et al.1989). Interestingly, polygenic analysis revealed that CUP2 also plays a role in acetic acid tolerance (Meijnen et al.2016) and a recent report has shown that Zygosaccharomyces bailii has only one HAA1/CUP2 ortholog which is involved both in acetic acid and copper tolerance (Palma et al.2017).
Recently, Gonzalez-Ramos et al. (2016) evolved a laboratory strain by serial microaerobic batch culture to obtain mutants with a superior acetic acid resistance phenotype. By analyzing whole-genome sequencing data, a common set of genes had acquired mutations during the evolutionary engineering. GIS4, SKS1, ADH3, ASG1, SAC6 and EUG1 had acquired SNPs in most of the evolved strains. The first four were confirmed by introducing them in a non-evolved strain and the mutations in GIS4, SKS1 and ADH3 had an additive effect. These genes had never been linked before to acetic acid tolerance by transcriptional profiling or QTL analysis (Li and Yuan, 2010; Mira et al. 2010; Ding et al.2013; Meijnen et al.2016). The successful application of these alleles remains to be done in an industrially relevant strain.
Other chemical inhibitors
A first class of chemical inhibitors present in lignocellulose hydrolysates are the furaldehydes, such as furfural and 5’-(hydroxymethyl)-2-furfural (HMF), which are conversion products of hexoses and pentoses, respectively. This conversion is mainly due to pretreatment methods applying high heat and high pressure. HMF is less inhibitory compared to furfural (Taherzadeh et al.2000). The main mechanism involved in tolerance to furaldehydes is that yeast converts these furaldehydes into less toxic compounds such as furfuryl alcohol and 2,5-bis-hydroxymethylfuran, respectively. This is most likely catalyzed by NAD(P)H-dependent (glycolytic) dehydrogenases (Modig, Liden and Taherzadeh 2002; Liu et al.2004). To achieve complete oxidation of 2.25g/L furfural in a hydrolysate, Hórvath et al. (2001) calculated that the cells require 2.5 times more ATP than in the absence of the furfural. In addition to competitive inhibition of alcohol dehydrogenase 1, furfural and HMF also have a direct allosteric inhibitory effect on the enzyme, resulting in a lower final ethanol titer.
Tolerance to furaldehydes has been linked to the expression of pentose phosphate pathway genes, and it was shown that overexpression of ZWF1, an oxidoreductase, enhanced tolerance to furfural, most likely due to an increase in cellular reducing power (Gorsich et al.2006). Cunha et al. (2015) confirmed the beneficial effect of ZWF1 and also linked overexpression of PRS3 and RPB4 to improved furaldehyde resistance. Many genes of the pleiotropic drug resistance (PDR) gene family have also been implicated and we refer the reader to the review of Liu for a comprehensive account (Liu 2011).
A second class of chemical inhibitors are the phenolic compounds derived from the lignin in lignocellulosic biomass. Lignin is a complex organic polymer of phenolic compounds, including acids, aldehydes and alcohols (Campbell and Sederoff 1996). The main mechanism of yeast tolerance is not due to export of the phenolic compounds, but, as for the furaldehydes, to in situ detoxification. Coniferyl aldehyde has been identified as the most toxic phenolic compound in the lignin fraction, and there seems to be a correlation between specific chemical classes and toxicity (Adeboye, Bettiga and Olsson 2014; Adeboye et al.2015). Lignin is very hydrophobic and binds to cellulose, preventing cellulase access for its hydrolysis. In addition, lignin also interacts with the cellulose-binding module of cellulases further reducing hydrolytic efficiency (Eriksson, Börjesson and Tjerneld 2002; Kumar et al.2012; Vermaas et al.2015). Moreover, genes that are involved in aromatic acid conversion, such as PAD1 and FDC1, have been shown to increase tolerance to furaldehydes and to enhance final ethanol yield (Larsson, Nilvebrant and Jönsson 2001; Mukai et al.2010). The validation of this approach in an industrial context, however, has not been reported.
Combined stress factors and intrinsic stress tolerance
Transcription of HSPs such as Hsp104 and Hsp12 as well as trehalose biosynthesis enzymes has in many cases been shown to be upregulated under ethanol stress. HSPs and trehalose not only have a protective function against ethanol stress, but also to many of the abovementioned stress factors (Wiemken 1990; Neves and François 1992; Mansure et al.1994; Ribeiro, Silva and Panek 1994; Lindquist and Kim 1996; Feder and Hofmann 1999). Recent research has revealed that at least under certain stress conditions it is not so much trehalose itself that exerts a protective function on the cell, but that the Tps1 protein itself is in some way involved (Petitjean et al.2015, 2016).
The general stress response is important because during the bioethanol production process multiple stress factors act together in different combinations depending on the timing in the fermentation, the process conditions and the type of biomass used. For instance, Woo et al. (2014) assessed the specific growth rate at increasing acetic acid levels at two temperatures, 30°C and 42°C, and also under combined ethanol and temperature stress, and they observed synergistically acting negative effects in both cases.
Metabolic stress
Production of bio-based chemicals requires a microorganism that can cope with a high level of metabolic stress. Since these chemicals are not natively produced by S. cerevisiae, or at least not in the high amounts required for an industrial production process, engineering of metabolic pathways and overexpression of recombinant enzymes generally leads to diverse stress factors, especially protein burden, possible redox imbalances and product inhibition.
Protein burden
Strong overexpression of recombinant proteins in order to create a superior yeast cell factory can cause a metabolic burden or protein burden on the cell. This can be caused by the high demand for energy, reducing power and amino acid building blocks for the production of the high levels of recombinant proteins, and/or competition between expressed proteins at the level of the protein biosynthetic machinery, and/or the secretion pathway. Another problem can be protein crowding in a confined cellular space (Srienc, Campbell and Bailey 1986; Sardonini and DiBiasio 1987; Gopal, Broad and Lloyd 1989; Janes et al.1990; Nevoigt 2008). As a result, the overexpression of proteins can cause in many cases an impaired specific growth rate, delayed fermentation start-up, reduced fermentation rate and/or yield, decreased respiration capacity or reduced biomass yield. The cost of protein production in bacteria is commonly attributed to protein translation, and ribosomal activity is considered to be a major limiting factor for the growth rate of the cells (Emilsson and Kurland 1990; Kurland 1992; Vind et al.1993; Scott et al.2010, 2014; Scott and Hwa 2011).
Only a limited amount of insight in the molecular mechanisms of protein burden in eukaryotic cells is available (Hauf, Zimmermann and Müller 2000; Lang, Murray and Botstein 2009). The concept of metabolic burden in S. cerevisiae was investigated and elaborated by Görgens et al. (2001). They compared S. cerevisiae strains expressing a heterologous xylanase II (XYN2) from Trichoderma reesei and quantified the metabolic effects of expression of plasmid-based constructs behind a glycolytic promoter (ADH2 or PGK1). Their research showed no significant burden on the cells when the heterologous gene is expressed from a multicopy plasmid. However, when the gene was expressed behind a glycolytic promoter, a reduction in maximum specific growth rate, biomass yield and specific glucose consumption was observed. They describe the metabolic effect of foreign gene expression as ‘disproportionally large with respect to the amount of heterologous protein produced’. The most obvious reasons for their findings are the increased energy demand and/or the competition for limiting amounts of transcription or translation factors, precursors and energy (Görgens et al.2001). However, alternative explanations such as negative interference of the foreign, heterologous protein or the artificial recombinant protein with cellular factors important for growth and/or fermentation cannot be ruled out. The effect of recombinant gene expression on transcription and protein translation in S. cerevisiae was studied by Kafri et al. (2016). They showed that the processes that limit protein production depend on the growth conditions, that ribosomal activity is not universally limiting in rapidly growing cells and that cells can adapt the abundance of endogenous proteins to meet at least to some extent the metabolic burden.
In Schizosaccharomyces pombe, recombinant protein production can be limited by lipid biosynthesis, TCA cycle activity and the supply of NADPH and ATP. Another important factor is the nutrient composition of the medium that can strongly influence protein secretion (Klein et al.2014). A similar effect was detected in S. cerevisiae, in which supplementation of the growth medium with amino acids improves growth and protein production (van Rensburg et al.2012). Furthermore, high gene copy numbers may strongly affect the metabolic activity of the cell (Lin et al.2013; Klein et al.2014). To determine the copy number limit for overexpression of a target gene, Makanae and co-workers developed a method named ‘genetic tug-of-war’ (gTOW). Using this genetic method, they concluded that the yeast cell is robust to a copy number increase by up to 100 in more than 80% of its protein-coding genes. Also, they identified 115 dosage-sensitive genes of which a significant number are genes involved in cytoskeletal organization and intracellular transport. This dosage sensitivity was suggested to be caused by protein burden and stoichiometric imbalances (Makanae et al.2013).
Redox factor imbalance
Another frequently encountered stress factor in cells modified by metabolic engineering is a disturbance in the balance of redox factors. The preference of enzymes for specific cofactors leads to a depletion and/or accumulation for instance of NAD(P)H. Hence, metabolic engineering of yeast requires appropriate modelling of redox homeostasis with correct prediction of cofactor usage (Nevoigt 2008).
The first efforts to engineer xylose fermentation into S. cerevisiae involved expression of xylose reductase (XYL1) and xylitol dehydrogenase (XYL2) of Pichia stipitis. This created a slow-growing strain with a low fermentation rate on xylose (Jeffries and Jin 2004). Since Xyl1 prefers NADPH over NADH and Xyl2 only uses NAD+ (Verduyn et al.1985; Rizzi et al.1989), it led to inability of recycling NADH and thus accumulation of NAD+ (Jeffries 2006).
Several approaches have been used to overcome redox factor imbalances. The modification of cofactor specificity is undertaken frequently in metabolic engineering, as was the case in pentose utilization with the identification of mutant xylose reductase enzymes that have a higher affinity for NADH than for NADPH (Jeppsson et al.2006; Watanabe et al. 2007a). Another approach was the engineering of xylitol dehydrogenase for NADP+ cofactor specificity (Watanabe et al. 2007b).
Recent research on 2,3-butanediol production was also confronted with the issue of redox imbalance. It was caused by deletion of the PDC genes, with the aim of abolishing ethanol production for enhancing the flux into the 2,3-butanediol production pathway (Kim et al.2013). In this case, the cofactor imbalance led to production of glycerol instead of 2,3-butanediol, because the disequilibrium was balanced by conversion of NADH to NAD+ (Remize et al.1999; Kim et al.2016). However, it could be corrected by expressing NADH oxidase from Lactococcus lactis (noxE), obtaining a maximal 2,3-butanediol yield (Kim and Hahn 2015; Kim et al.2015, 2016).
Another approach is the expression of a transhydrogenase that is natively absent in yeast (Bruinenberg et al.1985), catalyzing the following reaction: NADH + NADP+ → NAD+ + NADPH (Sauer et al. 2004b). Implementation of transhydrogenase activity can balance the disturbed redox state in metabolically engineered yeast as shown by Suga et al. (2013). They overexpressed three enzymes, i.e. Mae1, Mdh2 and Pyc2, that have a transhydrogenase-like activity in a xylose-fermenting S. cerevisiae strain, and this allowed proper control of the redox state. However, expression of a bacterial transhydrogenase in yeast resulted in a significant decrease in maximal specific growth rate, biomass and ethanol yield (Nissen et al.2001).
Toxic metabolites
Industrial-scale production of bulk and fine chemicals requires high titers of the end product (Stephanopoulos 2007). High concentrations of these chemicals are often toxic to the producing host cell (Keasling 2010). Imbalances introduced by suboptimal metabolic engineering often lead to accumulation of metabolic intermediates that may also be toxic to the cells. An example is the frequent accumulation of the toxic intermediate acetaldehyde in engineered yeast cell factories (Ng et al.2012; Bae, Kim and Hahn 2016). This cytotoxic compound, which is also produced in low quantities during alcoholic fermentation, diffuses poorly across the plasma membrane, which leads to intracellular accumulation and inhibition of sensitive reactions and enzymes (Aranda and del Olmo 2004).
One way to address this type of metabolic stress is through upregulation of efflux pumps (Kell et al.2015; Sheng and Feng 2015). Overexpression of membrane exporters can apparently enhance the yield of cytotoxic products. For instance, production of short branched-chain fatty acids is enhanced by overexpression of the native Pdr12 efflux pump (Yu et al.2016). Overexpression of the S. cerevisiae Snq2, Pdr5 and Pdr15 transporters was shown to increase tolerance against and secretion of alkanes (Ling et al.2013). Similarly, expressing the Abc2 and Abc3 membrane transporters of Yarrowia lipolytica in S. cerevisiae improved its tolerance to alkanes (Chen, Ling and Chang 2013).
Recent studies have explored another potential solution to address toxic effects of intermediary metabolites using the concept of ‘metabolons’. This strategy has actually been adapted from nature since biosynthetic enzymes are also sometimes physically associated in large complexes (Jorgensen et al.2005; Zhang et al.2006; Trantas, Panopoulos and Ververidis 2009; Singleton, Howard and Smirnoff 2014). This spatial localization does not only facilitate the transfer of intermediates between the consecutive enzymes of a pathway due to channeling of the metabolites, but it also prevents the diffusion of the intermediates away from the enzyme complex keeping their overall level far below a concentration that otherwise could become toxic for the cells (Siddiqui et al.2012). Minimizing the diffusion of toxic intermediates while stimulating their rapid conversion to less toxic constituents through sequestration within an enzyme complex might thus improve cell growth and especially the final yield of the product of interest (Srere 1987; Jorgensen et al.2005; Roze, Chanda and Linz 2011; Chen and Silver 2012).
STRAIN ENGINEERING FOR IMPROVING STRESS TOLERANCE
Saccharomyces cerevisiae provides excellent platform strains for conversion into cell factories. The species already possesses several traits that are required for cell factories, such as high fermentation rate, high propagation rate, high general robustness, high tolerance to alcohols and ease of genetic modification. These traits are even more pronounced in the many industrial strains that have been developed over the years, including those for bioethanol production that show excellent tolerance to most stress factors prevalent in first-generation bioethanol production.
Tolerance against these stress factors is also of great importance for creation of efficient yeast cell factories. Tolerance to specific stress factors can be enhanced through random genetic modification followed by selection or through targeted genetic modification based on previous knowledge of genetic determinants underlying tolerance to specific stress factors or general stress tolerance. The former can be achieved by random mutagenesis, genome shuffling, whole-genome hybridization or evolutionary adaptation, each time followed by selection of superior strains under a specific condition (Steensels et al.2014; David and Siewers 2015; Jullesson et al.2015). A disadvantage of all these approaches is that they can only be used for selective traits for which selection is possible based on growth rate or maintenance of viability. An advantage is that they generally lead to non-GMO strains that can be brought rapidly into industrial application.
Basic yeast research has produced a wealth of information on genetic determinants of tolerance to specific stress factors. The usefulness of this information has been limited by two major factors. The first is that most of this research has been performed with laboratory yeast strains which generally have much weaker tolerance to stress factors than industrial yeast strains. The second is that most information is available on genetic factors required for tolerance to stress factors and of which therefore the deletion reduces stress tolerance. Much less information is available on genetic factors that can be modified (by overexpression or by specific mutagenesis) to enhance tolerance to stress factors. As opposed to the random approaches, targeted genetic modification generally leads to GMO strains, either cisgenic (containing extra or modified species-own DNA only) or transgenic (containing heterologous DNA) organisms.
Stress tolerance is a complex, polygenic trait and there is little information on genetic determinants that can improve tolerance in industrial yeast strains to specific stress factors and even less to a mixture of stress factors. As a result, improvement of stress tolerance by non-directed engineering methodologies has been more successful up to now for development of superior industrial yeast strains used for bioethanol production compared to targeted genetic modification strategies (Cakar et al.2005; Gorsich et al.2006; Petersson et al.2006; Shi, Wang and Wang 2009; Hou 2010; Cakar et al.2012; Demeke et al.2013; Steensels et al.2014; Gonzalez-Ramos et al.2016).
Evolutionary engineering
Evolutionary engineering is a non-directed methodology to improve yeast strains for a specific trait by subjecting the cells to a continuous selective pressure. In repetitive batch cultivations or chemostat cultivation under selective pressure, genetic diversity is generated through spontaneous mutagenesis and the more fitter cells are continuously selected because of faster multiplication or better survival. The most adapted lineage(s) will finally dominate the entire culture after prolonged cultivation (Dykhuizen and Hartl 1983; Sauer 2001).
Evolutionary engineering has been applied to improve several industrially important traits in S. cerevisiae, such as substrate utilization, product formation and stress tolerance. Ethanol fermentation with different carbon sources was enhanced successfully. Rapid xylose utilization is one of the main requirements for yeast strains used in second-generation bioethanol production. This challenge has been addressed extensively using evolutionary engineering (Sonderegger and Sauer 2003; Kuyper et al.2005; van Maris et al.2007; Liu and Hu 2010; Zhou et al.2012; Demeke et al.2013). Other carbon sources for which utilization has been improved by evolutionary engineering include arabinose (Wisselink et al.2007; Sanchez et al.2010) and lactose (Guimaraes et al.2008). Furthermore, improvement of several stress tolerance characteristics has been achieved successfully by evolutionary engineering, such as freeze tolerance (Teunissen et al.2002), ethanol tolerance (Stanley et al. 2010b) and even multistress resistance (Cakar et al.2005).
It is highly relevant for industrial application to improve multiple traits at the same time by applying different selective pressures simultaneously in order to achieve the best possible combination of the individual adaptations. Wright et al. (2011) performed evolutionary engineering in medium with acetic acid and xylose to obtain a superior xylose-fermenting yeast strain under industrial conditions. Performing repetitive batch cultivations directly in lignocellulosic hydrolysates selects evolved strains that show increased tolerance to all inhibitors present in the hydrolysates (Tomas-Pejo et al.2010; Koppram, Albers and Olsson 2012).
Genome shuffling
Genome shuffling is extensive inbreeding of two or more yeast strains after which the strain(s) with the desirable trait is obtained by selection for faster growth or maintenance of higher viability under a specific condition of interest. Alternatively, the strains can also be mixed repeatedly by polyethylene glycol-mediated protoplast fusion. It is a whole-genome engineering technology that introduces many random changes at many different positions in the genome. Recombination of desirable mutant genes into a single strain can generate strongly improved phenotypes (Nicolaou Gaida and Papoutsakis 2010). In S. cerevisiae, this technique has been applied successfully to enhance tolerance to specific stress factors. Shi, Wang and Wang (2009) enhanced thermotolerance, ethanol productivity and ethanol tolerance using genome shuffling. Their best performing strain was claimed to grow at up to 55°C on plates. Moreover, it utilized 20% (w/v) glucose at 45°C –48°C and produced 9.95% (w/v) ethanol while it tolerated up to 25% (v/v) ethanol.
The efficiency of protoplast fusion is crucial for efficient genome shuffling using this method. Eukaryotic hybrids are unstable due to diversion of the genetic background of the parents (Giudici et al.2005). To address this issue, researchers have attempted to make genome shuffling more efficient in several ways. For instance, the number of potentially useful mutations was increased by ethyl methane sulfonate (EMS) mutagenesis followed by sexual and asexual reproduction instead of protoplast fusion (Hou 2009). This methodology was applied successfully to improve ethanol productivity (Hou 2010) as well as stress tolerance (Pinel et al.2011) and fermentation performance (Zheng et al. 2011a). In other research, acetic acid tolerance in S. cerevisiae was enhanced using drug resistance marker-aided genome shuffling, which was proven to be advantageous for highly efficient selection of the genome shuffled mutants (Zheng et al. 2011b). Demeke et al. (2013) developed an industrial D-xylose fermenting strain through genome shuffling by sporulation and mass mating of an EMS mutagenized segregant and its parent, followed by evolutionary adaptation.
The genome shuffling method has also been modified to counter other disadvantages of protoplast fusion, such as time-consuming protoplast preparation, fusant regeneration and fusant instability (Zhang and Geng 2012). Genomic DNA extraction from one parent followed by its transfer into the other parent also allowed recombination of the two genomes. To allow a second round of shuffling, the strain obtained can be transformed again with the whole genome of the first parent. Such a modified interspecies genome shuffling methodology was used successfully to construct a xylose-fermenting S. cerevisiae strain through transformation with the whole genome of Pichia stipitis (Zhang and Geng 2012).
CRISPR-Cas9
Genome editing is highly dependent on the efficacy of the engineering methodology used. In recent years, a very precise genetic targeting and modification technology, that is based on clustered regularly interspaced palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas), has gained widespread application in many species because of its high efficacy and flexibility (Doudna and Charpentier 2014). Since this is a targeted modification strategy, it requires knowledge of the genes that are important for a specific trait of interest such as genes determining tolerance to specific stress factors. Determining the causative elements is thus essential prior to improving the yeast. This can be done by previously described methodologies such as pooled-segregant whole-genome sequence analysis (Segre, Murray and Leu 2006; Swinnen, Thevelein and Nevoigt 2012). This technique was applied successfully to identify causative genes for several industrially important traits, such as high ethanol tolerance (Swinnen et al. 2012; Pais et al.2013), acetic acid tolerance (Meijnen et al.2016), low glycerol production (Hubmann et al.2013) and thermotolerance (Yang et al.2013).
Genome engineering in S. cerevisiae reached a new level of efficiency with CRISPR-Cas9, as described by DiCarlo et al. (2013), and also strongly facilitated targeted genome engineering in industrial yeast strains (Stovicek, Borodina and Forster 2015). This technology can be applied for introducing both single and multiple marker-free gene knockouts (Bao et al.2015; Mans et al.2015), as was the case for establishing high mevalonate production in yeast (Jakociunas et al. 2015a). Besides gene knockouts, successful CRISPR-Cas9-mediated gene integration can be achieved, which is often required in development of yeast cell factories for bio-based chemical production (Jakociunas, Jensen and Keasling 2016). Marker-free integration of single or multiple genes, or even entire metabolic pathways, has become possible in a single step with the application of CRISPR-Cas9 combined with homologous recombination. Evaluation of a gene library by direct genomic insertion in a diploid strain in order to identify the most active variants of a cellodextrin transporter for increased cellobiose utilization was achieved in a single step (Ryan et al.2014).
Adaptation of the original CRISPR-Cas9 method by combination with in vivo assembly using homologous recombination (CasEMBLR) makes it more versatile (Jakociunas et al. 2015b). The applicability of this approach was validated by introduction of the carotenoid pathway through insertion of 15 DNA fragments in multiple loci and through creating a tyrosine producing strain by simultaneously knocking out two genes and integrating 10 DNA fragments (Jakociunas et al. 2015b). This methodology of combining CRISPR-Cas9 with in vivo assembly has been used for introduction of several traits in yeast. Horwitz et al. (2015) chromosomally integrated an entire 11-gene pathway for muconic acid production, and Tsai et al. (2015) constructed a xylose-fermenting strain in this way.
Another recent study designed specific guide RNA sequences to target multiple delta sites (Di-CRISPR) (Shi et al.2016). This allows simultaneous integration of multiple copies in different positions in the yeast genome. The authors successfully integrated 18 copies of a 24 kb combined xylose utilization and (R,R)-2,3-butanediol (BDO) production pathway with a high efficiency in a single marker-free step in the delta elements, thus creating a yeast strain that produced BDO directly from xylose (Shi et al.2016). This approach is of great interest for integrating multiple copies of a gene or a pathway in a single step into the genome.
Global transcription machinery engineering
A relatively new technology to improve important phenotypes in cell factories is global transcription machinery engineering (gTME). This non-directed approach is based on mutagenesis of transcription factors and/or cofactors (Nevoigt 2008). Altering key regulatory proteins generates a new transcription profile by reprogramming gene transcription. This was first accomplished by Alper and co-workers (Alper et al.2006; Ding et al.2009), who created a strain with increased ethanol tolerance caused by an altered TATA-binding protein Spt15. Three mutations in SPT15 (F177S, Y195H and K218R) improved the mutant's performance compared to the control in the presence of high ethanol concentrations. These findings were challenged by Baerends et al. (2009) who could not observe the same effect in leucine prototrophic strains and ascribed the original effects on ethanol tolerance to alleviation of leucine auxotrophy. On the other hand, Yang et al. (2011) used the same approach with the Spt15 transcription factor and also successfully demonstrated improved ethanol tolerance in rich medium with specific mutant alleles. However, they also used only leucine auxotrophic strains.
Besides enhancement of ethanol tolerance, gTME has also been successfully applied in S. cerevisiae to improve xylose utilization (Liu et al.2010) and fermentation in corn cob hydrolysates (Liu et al.2011), and to enhance co-fermentation of xylose and glucose (Liu et al.2008). This implies that gTME may be a promising technique for strain improvement but that more positive results especially with industrial strains and industrially relevant conditions are required.
INDUSTRIAL USE OF ALTERNATIVE STRESS-TOLERANT NON-CONVENTIONAL YEAST SPECIES
Saccharomyces cerevisiae is still the preferred yeast species in many industrial applications, but there is a plethora of other yeast species that have some very desirable traits, and the use of some of these as cell factories for production of bio-based compounds has been investigated. However, very often they lack the general robustness that S. cerevisiae has acquired during its domestication and extensive use in industry. Another major shortcoming is their requirement for aerobic conditions, whereas S. cerevisiae is able to accomplish complete fermentations in the anaerobic conditions prevailing in large non-aerated fermentors.
Several species of the genus Zygosaccharomyces possess extreme tolerance to specific stress factors. Zygosaccharomyces rouxii and Z. bailii are both well-known food spoilage yeasts due to their extreme osmotolerance and weak acid tolerance, respectively (Martorell et al.2007). Production of miso makes use of the very osmotolerant character of Z. rouxii to produce furanones, which give miso its typical taste (Hayashida, Nishimura and Slaughter 1998). Zygosaccharomyces rouxii has also been used as cell factory for recombinant protein production. Although similar titers as in S. cerevisiae have been obtained for some proteins, industrial production has never been achieved, possibly due to the limited knowledge and experience with Z. rouxii in industry (Ogawa et al.1990). More recently, identification of the mechanisms underlying its very high osmotolerance has become of interest in order to ameliorate osmotolerance in established industrial microorganisms (Mattanovich et al.2012).
Another very osmotolerant yeast species is Debaryomyces hansenii. It can tolerate salt concentrations over 4.0 M NaCl, while S. cerevisiae is not able to grow in the presence of over 2.0 M NaCl (ŌNishi 1963). This feature would give it an advantage in fermentation media containing high salt levels, e.g. due to the pretreatment process, recirculation of water, cleaning practices, use of feedstocks rich in salt or use of seawater. Despite being one of the best xylitol producing yeast species, industrial implementation has been hampered by its dependency on oxygen (Breuer and Harms 2006). As with Z. rouxii, research has been focused on identifying genes responsible for the high salt tolerance of D. hansenii in order to improve industrial strains of established yeast species. In this regard, an S. cerevisiae lab strain displayed improved salt tolerance by expression of genes from D. hansenii (Prista et al.2002).
Zygosaccharomyces bailii has been more extensively investigated as a cell factory because of its high tolerance to weak acids. Many of these acids are of interest as low value bulk chemicals (e.g.s acetic acid, L-ascorbic acid, succinic acid, and so on), but are only tolerated in low concentrations by most microorganisms. L-Ascorbic acid production was engineered both in S. cerevisiae and Z. bailii, obtaining a much higher titer using Z. bailii (Sauer et al. 2004a). In addition, the molecular tools developed so far for Z. bailii are more advanced than for Z. rouxii (Branduardi, Dato and Porro 2014).
Two yeast species relatively closely related to S. cerevisiae can grow up to at least 49°C: Hansenula polymorpha and Kluyveromyces marxianus (Banat, Nigam and Marchant 1992; Reinders et al.1999). Hansenula polymorpha is a methylotrophic yeast that is mostly used as a model for investigation of peroxisome function and nitrate assimilation (Kunze, Kang and Gellissen 2009). Its ability to grow on xylose and methanol combined with its high thermotolerance has also created an interest to use it as a cell factory. Examples of recombinant protein production with H. polymorpha include the production of phytase and human IFNα-2a at lab scale (Mayer et al.1998; Degelmann et al.2002) and human serum albumin at pilot scale (Heo et al.2003). Despite these successes, the H. polymorpha expression systems and the long fermentation times are major drawbacks for the organism to become routinely used in industrial production.
Unlike D. hansenii, K. marxianus has a longer track record in industry. Its GRAS status (generally recognized as safe), broad substrate range and thermotolerance make it an attractive host for recombinant protein production. Whereas S. cerevisiae has only limited protein secretory capacity, K. marxianus has become a natural secretor during evolution for multiple enzymes, including inulinase, polygalacturonases and ß-glucosidase (Fonseca et al.2008). Schwan, Cooper and Wheals (1997) reported that in K. marxianus strain CCT3172 endopolygalacturonase was the only secreted enzyme, making it preferred over other species/strains due to lower downstream processing costs. In spite of its promising characteristics, most K. marxianus strains require sufficient aeration and tend to overglycosylate (recombinant) proteins (Hensing et al.1994). Furthermore, high sugar concentrations inhibit ethanol production in K. marxianus, making this a less attractive host for bio-based fuel production (Margaritis and Bajpaj 1983).
CONCLUSIONS
This review has discussed the main stress factors encountered by yeast cell factories used for production of bioethanol and bio-based chemicals and also how to engineer the cell factories for increased stress tolerance. Yeast cell factories are subject to a variety of stress factors during first- and second-generation fermentation processes. The industry is in dire need of microorganisms that can cope with the multitude of stress factors that can occur.
A variety of yeast species possess specific stress tolerance characteristics that are of interest to the industry but lack other desirable traits. Saccharomyces cerevisiae is still considered to be one of the most robust and versatile microorganisms. Its long-standing track record in the fermentation industry makes it an obvious choice for further improvement of its stress tolerance properties to make it even more suitable for use in biofuel and bio-based chemical production with difficult feedstocks. However, S. cerevisiae is lacking the capacity for utilization or production of certain compounds, it can also make undesirable post-translational modifications in protein production and has lower tolerance to certain stress factors than other species. Therefore, other microorganisms could be favored for specific industrial production processes.
Most information on the molecular-genetic basis of stress tolerance has been gained with laboratory yeast strains. This information is often not applicable to industrial yeast strains. Hence, more research is needed on the genetic basis of stress tolerance characteristics in industrial and natural yeast strains. These strains contain interesting alleles for targeted strain improvement while minimizing the risks of side effects on other industrially important traits.
Most research has also concentrated on genetic elements that are required to maintain stress tolerance and much less on genetic modifications that can improve stress tolerance. Since industrial yeast strains are generally much more robust and stress tolerant than laboratory yeast strains, more research on genetic factors important for stress tolerance and especially enhancement of stress tolerance in industrial yeast strains is required.
Genetic modifications that improve stress tolerance in yeast can compromise other properties that are important for industrial application, such as fermentation performance, propagation rate and tolerance to drying and storage conditions. Hence, it is important to evaluate newly constructed yeast strains under as many conditions as possible that are relevant in the industry.
The recent advent of highly powerful and precise genome engineering technologies, especially the CRISPR-Cas9 technology, has enabled much faster engineering of industrial yeast strains and will hopefully stimulate the development of highly productive cell factories for various applications in the production of biofuels and bio-based chemicals.
Conflict of interest.None declared.
REFERENCES
- Abdel-Banat BM, Hoshida H, Ano A et al. . High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl Microbiol Biot 2010;85:861–7. [DOI] [PubMed] [Google Scholar]
- Abdullah N, Nawawi A, Othman I. Fungal spoilage of starch-based foods in relation to its water activity (aw). J Stored Prod Res 2000;36:47–54. [Google Scholar]
- Adeboye PT, Bettiga M, Aldaeus F et al. . Catabolism of coniferyl aldehyde, ferulic acid and p-coumaric acid by Saccharomyces cerevisiae yields less toxic products. Microb Cell Fact 2015;14:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeboye PT, Bettiga M, Olsson L. The chemical nature of phenolic compounds determines their toxicity and induces distinct physiological responses in Saccharomyces cerevisiae in lignocellulose hydrolysates. AMB Express 2014;4:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera F, Peinado RA, Millan C et al. . Relationship between ethanol tolerance, H+ -ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains. Int J Food Microbiol 2006;110:34–42. [DOI] [PubMed] [Google Scholar]
- Alexandre H, Ansanay-Galeote V, Dequin S et al. . Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett 2001;498:98–103. [DOI] [PubMed] [Google Scholar]
- Alper H, Moxley J, Nevoigt E et al. . Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 2006;314:1565–8. [DOI] [PubMed] [Google Scholar]
- Aranda A, del Olmo Ml. Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl Environ Microb 2004;70:1913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SJ, Kim S, Hahn JS. Efficient production of acetoin in Saccharomyces cerevisiae by disruption of 2,3-butanediol dehydrogenase and expression of NADH oxidase. Sci Rep 2016;6:27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends RJ, Qiu JL, Rasmussen S et al. . Impaired uptake and/or utilization of leucine by Saccharomyces cerevisiae is suppressed by the SPT15-300 allele of the TATA-binding protein gene. Appl Environ Microb 2009;75:6055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banat IM, Nigam P, Marchant R. Isolation of thermotolerant, fermentative yeasts growing at 52°C and producing ethanol at 45°C and 50°C. World J Microb Biot 1992;8:259–63. [DOI] [PubMed] [Google Scholar]
- Bao Z, Xiao H, Liang J et al. . Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol 2015;4:585–94. [DOI] [PubMed] [Google Scholar]
- Basso LC, de Amorim HV, de Oliveira AJ et al. . Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res 2008;8:1155–63. [DOI] [PubMed] [Google Scholar]
- Bautista-Gallego J, Arroyo-Lopez FN, Durán-Quintana MC et al. . Individual effects of sodium, potassium, calcium, and magnesium chloride salts on Lactobacillus pentosus and Saccharomyces cerevisiae growth. J Food Protect 2008;71:1412–21. [DOI] [PubMed] [Google Scholar]
- Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels -Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 2012;23:631–40. [DOI] [PubMed] [Google Scholar]
- Bellissimi E, van Dijken JP, Pronk JT et al. . Effects of acetic acid on the kinetics of xylose fermentation by an engineered, xylose-isomerase-based Saccharomyces cerevisiae strain. FEMS Yeast Res 2009;9:358–64. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Vandenberghe LPS, Soccol CR et al. . First generation bioethanol. In: Green Fuels Technology, Springer International Publishing, Switzerland: 2016, 175–212. [Google Scholar]
- Branduardi P, Dato L, Porro D. Molecular tools and protocols for engineering the acid-tolerant yeast Zygosaccharomyces bailii as a potential cell factory. Methods Mol Biol 2014;1152:63–85. [DOI] [PubMed] [Google Scholar]
- Breuer U, Harms H. Debaryomyces hansenii—an extremophilic yeast with biotechnological potential. Yeast 2006;23:415–37. [DOI] [PubMed] [Google Scholar]
- Brown AD. Microbial water stress. Bacteriol Rev 1976;40:803–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinenberg PM, Jonker R, van Dijken JP et al. . Utilization of formate as an additional energy source by glucose-limited chemostat cultures of Candida utilis CBS 621 and Saccharomyces cerevisiae CBS 8066. Arch Microbiol 1985;142:302–6. [Google Scholar]
- Buchman C, Skroch P, Welch J et al. . The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol Cell Biol 1989;9:\4091–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakar ZP, Seker UO, Tamerler C et al. . Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res 2005;5:569–78. [DOI] [PubMed] [Google Scholar]
- Cakar ZP, Turanli-Yildiz B, Alkim C et al. . Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res 2012;12:171–82. [DOI] [PubMed] [Google Scholar]
- Campbell MM, Sederoff RR. Variation in lignin content and composition. Plant Physiol 1996;110:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright CP, Juroszek J-R, Beavan MJ et al. . Ethanol dissipates the proton-motive force across the plasma membrane of Saccharomyces cerevisiae. J Gen Microbiol 1986;132:369–77. [Google Scholar]
- Cartwright P, Veazey FJ, Rose AH. Effect of ethanol on activity of the plasma membrane in, and accumulation of glycine by, Saccharomyces cerevisiae.pdf. J Gen Microbiol 1987;133:857–65. [DOI] [PubMed] [Google Scholar]
- Casey E, Mosier NS, Adamec J et al. . Effect of salts on the co-fermentation of glucose and xylose by a genetically engineered strain of Saccharomyces cerevisiae. Biotechnol Biofuels 2013;6:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspeta L, Chen Y, Ghiaci P et al. . Altered sterol composition renders yeast thermotolerant. Science 2014;346:75–8. [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS et al. . Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 2001;12:323–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Rodriguez MF, Mosqueira-Salazar KJ, Ensinas AV et al. . Water reuse and recycling according to stream qualities in sugar–ethanol plants. Energy Sustain Dev 2013;17:546–54. [Google Scholar]
- Chen AH, Silver PA. Designing biological compartmentalization. Trends Cell Biol 2012;22:662–70. [DOI] [PubMed] [Google Scholar]
- Chen B, Ling H, Chang MW. Transporter engineering for improved tolerance against alkane biofuels in Saccharomyces cerevisiae. Biotechnol Biofuels 2013;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Z, Arneborg N. Saccharomyces cerevisiae strains with different degrees of ethanol tolerance exhibit different adaptive responses to produced ethanol. J Ind Microbiol Biot 2000;24:75–8. [Google Scholar]
- Choudhary J, Singh S, Nain L. Thermotolerant fermenting yeasts for simultaneous saccharification fermentation of lignocellulosic biomass. Electronic J Biotechn 2016;21:82–92. [Google Scholar]
- Chung H, Yang JE, Ha JY et al. . Bio-based production of monomers and polymers by metabolically engineered microorganisms. Curr Opin Biotechnol 2015;36:73–84. [DOI] [PubMed] [Google Scholar]
- Cray JA, Stevenson A, Ball P et al. . Chaotropicity: a key factor in product tolerance of biofuel-producing microorganisms. Curr Opin Biotechnol 2015;33:228–59. [DOI] [PubMed] [Google Scholar]
- Cunha JT, Aguiar TQ, Romani A et al. . Contribution of PRS3, RPB4 and ZWF1 to the resistance of industrial Saccharomyces cerevisiae CCUG53310 and PE-2 strains to lignocellulosic hydrolysate-derived inhibitors. Bioresource Technol 2015;191:7–16. [DOI] [PubMed] [Google Scholar]
- David F, Siewers V. Advances in yeast genome engineering. FEMS Yeast Res 2015;15:1–14. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Hottiger T, Dominguez J et al. . The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant . Eur J Biochem 1994;219:179–86. [DOI] [PubMed] [Google Scholar]
- Degelmann A, Müller F, Sieber H et al. . Strain and process development for the production of human cytokines in Hansenula polymorpha. FEMS Yeast Res 2002;2:349–61. [DOI] [PubMed] [Google Scholar]
- Demeke M, Dietz H, Li Y et al. . Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 2013;6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P et al. . Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 2013;41:4336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Bierma J, Smith MR et al. . Acetic acid inhibits nutrient uptake in Saccharomyces cerevisiae: auxotrophy confounds the use of yeast deletion libraries for strain improvement. Appl Microbiol Biot 2013;97:7405–16. [DOI] [PubMed] [Google Scholar]
- Ding J, Huang X, Zhang L et al. . Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl Microbiol Biot 2009;85:253–63. [DOI] [PubMed] [Google Scholar]
- Dinh TN, Nagahisa K, Yoshikawa K et al. . Analysis of adaptation to high ethanol concentration in Saccharomyces cerevisiae using DNA microarray. Bioprocess Biosyst Eng 2009;32:681–8. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- Du B, Sharma LN, Becker C et al. . Effect of varying feedstock-pretreatment chemistry combinations on the formation and accumulation of potentially inhibitory degradation products in biomass hydrolysates. Biotechnol Bioeng 2010;107:430–40. [DOI] [PubMed] [Google Scholar]
- Dykhuizen DE, Hartl DL. Selection in chemostats. Microbiol Rev 1983;47:150–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Kurland CG. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J 1990;9:4359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus DJ, van der Merwe GK, van Vuuren HJJ. Genome-wide expression analyses: Metabolic adaptation to high sugar stress. FEMS Yeast Res 2003;3:375–99. [DOI] [PubMed] [Google Scholar]
- Eraso P, Gancedo C. Activation of yeast plasma membrane ATPase by acid pH during growth. FEBS Lett 1987;224:187–92. [DOI] [PubMed] [Google Scholar]
- Eriksson T, Börjesson J, Tjerneld F. Mechanism of surfactant effect in enzymatic hydrolysis of lignocellulose. Enzyme Microb Technol 2002;31:353–64. [Google Scholar]
- Fan Z, South C, Lyford K et al. . Conversion of paper sludge to ethanol in a semicontinuous solids-fed reactor. Bioprocess Biosyst Eng 2003;26:93–101. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat shock proteins, molecular chaperones, and stress response: evolutionary and ecological physiology. Ann Rev Physiol 1999;61:243–82. [DOI] [PubMed] [Google Scholar]
- Fernandes AR, Mira NP, Vargas RC et al. . Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Bioph Res Co 2005;337:95–103. [DOI] [PubMed] [Google Scholar]
- Fonseca GG, Heinzle E, Wittmann C et al. . The yeast Kluyveromyces marxianus and its biotechnological potential. Appl Microbiol Biot 2008;79:339–54. [DOI] [PubMed] [Google Scholar]
- Garre E, Raginel F, Palacios A et al. . Oxidative stress responses and lipid peroxidation damage are induced during dehydration in the production of dry active wine yeasts. Int J Food Microbiol 2010;136:295–303. [DOI] [PubMed] [Google Scholar]
- Giudici P, Solieri L, Pulvirenti AM et al. . Strategies and perspectives for genetic improvement of wine yeasts. Appl Microbiol Biot 2005;66:622–8. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ramos D, Gorter de Vries AR, Grijseels SS et al. . A new laboratory evolution approach to select for constitutive acetic acid tolerance in Saccharomyces cerevisiae and identification of causal mutations. Biotechnol Biofuels 2016;9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal CV, Broad D, Lloyd D. Bioenergetic consequences of protein overexpression in Saccharomyces cerevisiae. Appl Microbiol Biot 1989;30:160–5. [Google Scholar]
- Görgens JF, van Zyl WH, Knoetze JH et al. . The metabolic burden of the PGK1 and ADH2 promoter systems for heterologous xylanase production by Saccharomyces cerevisiae in defined medium Biotechnol Bioeng 2001;73:238–45. [DOI] [PubMed] [Google Scholar]
- Gorsich SW, Dien BS, Nichols NN et al. . Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biot 2006;71:339–49. [DOI] [PubMed] [Google Scholar]
- Graves T, Narendranath NV, Dawson K et al. . Effect of pH and lactic or acetic acid on ethanol productivity by Saccharomyces cerevisiae in corn mash. J Ind Microbiol Biotechnol 2006;33:469–74. [DOI] [PubMed] [Google Scholar]
- Guimaraes PM, Francois J, Parrou JL et al. . Adaptive evolution of a lactose-consuming Saccharomyces cerevisiae recombinant. Appl Environ Microb 2008;74:1748–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldfeldt LU, Arneborg N. Measurement of the effects of acetic acid and extracellular pH on intracellular pH of nonfermenting, individual Saccharomyces cerevisiae cells by fluorescence microscopy. Appl Environ Microb 1997;64:530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsworth JE, Nomura Y, Iwahara M. Ethanol-induced water stress and fungal growth. J Ferment Bioeng 1998;86:451–6. [Google Scholar]
- Hamelinck CN, Hooijdonk GV, Faaij APC. Ethanol from lignocellulosic biomass: techno-economic performance in short-, middle- and long-term. Biomass Bioenergy 2005;28:384–410. [Google Scholar]
- Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol 2006;46:215–34. [DOI] [PubMed] [Google Scholar]
- Hauf J, Zimmermann FK, Müller S. Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae. Enzyme Microb Technol 2000;26:688–98. [DOI] [PubMed] [Google Scholar]
- Hayashida Y, Nishimura K, Slaughter JC. The importance of the furanones HDMF and HEMF in the flavour profile of Japanese barley miso and their production during fermentation. J Sci Food Agr 1998;78:88–94. [Google Scholar]
- Henderson CM, Block DE. Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomyces cerevisiae. Appl Environ Microb 2014;80:2966–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensing M, Vrouwenvelder H, Hellinga C et al. . Production of extracellular inulinase in high-cell-density fed-batch cultures of Kluyveromyces marxianus. Appl Microbiol Biot 1994;42:516–21. [Google Scholar]
- Heo J, Hong W, Cho E et al. . Properties of the Hansenula polymorpha-derived constitutive promoter, assessed using an HSA reporter gene. FEMS Yeast Res 2003;4:175–84. [DOI] [PubMed] [Google Scholar]
- Hirasawa T, Yoshikawa K, Nakakura Y et al. . Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J Biotechnol 2007;131:34–44. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol R 2002;66:300–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Methods Enzymol 2007;428:29–45. [DOI] [PubMed] [Google Scholar]
- Horvat J, Klaic B, Metelko B et al. . Mechanism of levulinic acid formation. Tetrahedron Lett 1985;262111–4. [Google Scholar]
- Hórvath IS, Taherzadeh MJ, Niklasson C et al. . Effects of furfural on anaerobic continuous cultivation of Saccharomyces cerevisiae. Biotechnol Bioeng 2001;75:540–9. [DOI] [PubMed] [Google Scholar]
- Horwitz AA, Walter JM, Schubert MG et al. . Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst 2015;1:88–96. [DOI] [PubMed] [Google Scholar]
- Hou L. Novel methods of genome shuffling in Saccharomyces cerevisiae. Biotechnol Lett 2009;31:671–7. [DOI] [PubMed] [Google Scholar]
- Hou L. Improved production of ethanol by novel genome shuffling in Saccharomyces cerevisiae. Appl Biochem Biotech 2010;160:1084–93. [DOI] [PubMed] [Google Scholar]
- Hubmann G, Mathé L, Foulquie-Moreno MR et al. . Identification of multiple interacting alleles conferring low glycerol and high ethanol yield in Saccharomyces cerevisiae ethanolic fermentation. Biotechnol Biofuels 2013;6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffer S, Clark ME, Ning JC et al. . Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl Environ Microb 2011;77:6400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeas JI, Jimenez J. Mitochondrial DNA loss caused by ethanol in Saccharomyces flor yeasts. Appl Environ Microb 1997;63:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Ohno T. The relationship between viability and intracellular pH in the yeast Saccharomyces cerevisiae. Appl Environ Microb 1995;61:3604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakociunas T, Bonde I, Herrgard M et al. . Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 2015a;28:213–22. [DOI] [PubMed] [Google Scholar]
- Jakociunas T, Jensen MK, Keasling JD. CRISPR/Cas9 advances engineering of microbial cell factories. Metab Eng 2016;34: 44–59. [DOI] [PubMed] [Google Scholar]
- Jakociunas T, Rajkumar AS, Zhang J et al. . CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth Biol 2015b;4:1226–34. [DOI] [PubMed] [Google Scholar]
- Janes M, Meyhack B, Zimmermann W et al. . The influence of GAP promoter variants on hirudin production, average plasmid copy number and cell growth in Saccharomyces cerevisiae. Curr Genet 1990;18:97–103. [DOI] [PubMed] [Google Scholar]
- Jeffries TW. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 2006;17:320–6. [DOI] [PubMed] [Google Scholar]
- Jeffries TW, Jin YS. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biot 2004;63:495–509. [DOI] [PubMed] [Google Scholar]
- Jeppsson M, Bengtsson O, Franke K et al. . The expression of a Pichia stipitis xylose reductase mutant with higher K(M) for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 2006;93:665–73. [DOI] [PubMed] [Google Scholar]
- Jimenez J, Benítez T. Adaptation of yeast cell membranes to ethanol. Appl Environ Microb 1987;53:1196–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Dunlap PE, McBride SJ et al. . Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet 2008;4:e1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen K, Rasmussen AV, Morant M et al. . Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol 2005;8:280–91. [DOI] [PubMed] [Google Scholar]
- Jullesson D, David F, Pfleger B et al. . Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol Adv 2015;33:1395–402. [DOI] [PubMed] [Google Scholar]
- Kafri M, Metzl-Raz E, Jona G et al. . The cost of protein production. Cell Rep 2016;14:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaino T, Takagi H. Gene expression profiles and intracellular contents of stress protectants in Saccharomyces cerevisiae under ethanol and sorbitol stresses. Appl Microbiol Biot 2008;79:273–83. [DOI] [PubMed] [Google Scholar]
- Kampranis SC, Makris AM. Developing a yeast cell factory for the production of terpenoids. Comput Struct Biotechnol J 2012;3:e201210006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal JR, Kakkar VK, Malik NS et al. . Chemical composition of sugarcane bagasse and filter-mud and the effect of source and alkali treatment on the in vitro dry matter digestibility of bagasse. Ind J Dairy Sci 1981;34:458–62. [Google Scholar]
- Kawaguchi H, Hasunuma T, Ogino C et al. . Bioprocessing of bio-based chemicals produced from lignocellulosic feedstocks. Curr Opin Biotechnol 2016;42:30–9. [DOI] [PubMed] [Google Scholar]
- Kawahata M, Masaki K, Fujii T et al. . Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res 2006;6:924–36. [DOI] [PubMed] [Google Scholar]
- Keasling JD. Manufacturing molecules through metabolic engineering. Science 2010;330:1355–8. [DOI] [PubMed] [Google Scholar]
- Kell DB, Swainston N, Pir P et al. . Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol 2015;33:237–46. [DOI] [PubMed] [Google Scholar]
- Kim JW, Kim J, Seo SO et al. . Enhanced production of 2,3-butanediol by engineered Saccharomyces cerevisiae through fine-tuning of pyruvate decarboxylase and NADH oxidase activities. Biotechnol Biofuels 2016;9:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Seo SO, Zhang GC et al. . Expression of Lactococcus lactis NADH oxidase increases 2,3-butanediol production in Pdc-deficient Saccharomyces cerevisiae. Bioresource Technol 2015;191:512–9. [DOI] [PubMed] [Google Scholar]
- Kim S, Hahn JS. Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng 2015;31:94–101. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Seo SO, Jin YS et al. . Production of 2,3-butanediol by engineered Saccharomyces cerevisiae. Bioresource Technol 2013;146:274–81. [DOI] [PubMed] [Google Scholar]
- Klein T, Lange S, Wilhelm N et al. . Overcoming the metabolic burden of protein secretion in Schizosaccharomyces pombe–a quantitative approach using 13C-based metabolic flux analysis. Metab Eng 2014;21:34–45. [DOI] [PubMed] [Google Scholar]
- Koppram R, Albers E, Olsson L. Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol Biofuels 2012;5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen JB, Felby C, Jorgensen H. Yield-determining factors in high-solids enzymatic hydrolysis of lignocellulose. Biotechnol Biofuels 2009;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Arantes V, Chandra R et al. . The lignin present in steam pretreated softwood binds enzymes and limits cellulose accessibility. Bioresource Technol 2012;103:201–8. [DOI] [PubMed] [Google Scholar]
- Kunze G, Kang HA, Gellissen G. Hansenula polymorpha (Pichia angusta): biology and applications. In: Satyanarayana T, Kunze G (eds). Yeast Biotechnology: Diversity and Applications. Berlin, Heidelberg: Springer Science + Business Media B.V., 2009; 47–64. [Google Scholar]
- Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet 1992;26:29–50. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Toirkens MJ, Diderich JA et al. . Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res 2005;5:925–34. [DOI] [PubMed] [Google Scholar]
- Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness trade-off in yeast. P Natl Acad Sci USA 2009;106:5755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson S, Nilvebrant NO, Jönsson L. Effect of overexpression of Saccharomyces cerevisiae Pad1p on the resistance to phenylacrylic acids and lignocellulose hydrolysates under aerobic and oxygen-limited conditions. Appl Microbiol Biot 2001;57:167–74. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Elkon IM, McGee MA et al. . Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 2010;186:1197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BZ, Yuan YJ. Transcriptome shifts in response to furfural and acetic acid in Saccharomyces cerevisiae. Appl Microbiol Biot 2010;86:1915–24. [DOI] [PubMed] [Google Scholar]
- Limayem A, Ricke SC. Lignocellulosic biomass for bioethanol production: current perspectives, potential issues and future prospects. Prog Energ Combust 2012;38:449–67. [Google Scholar]
- Lin XQ, Liang SL, Han SY et al. . Quantitative iTRAQ LC-MS/MS proteomics reveals the cellular response to heterologous protein overexpression and the regulation of HAC1 in Pichia pastoris. J Proteomics 2013;91:58–72. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. P Natl Acad Sci USA 1996;93:5301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Chen B, Kang A et al. . Transcriptome response to alkane biofuels in Saccharomyces cerevisiae- identification of efflux pumps involved in alkane tolerance. Biotechnol Biofuels 2013;6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E, Hu Y. Construction of a xylose-fermenting Saccharomyces cerevisiae strain by combined approaches of genetic engineering, chemical mutagenesis and evolutionary adaptation. Biochem Eng J 2010;48:204–10. [Google Scholar]
- Liu H, Liu K, Yan M et al. . gTME for improved adaptation of Saccharomyces cerevisiae to corn cob acid hydrolysate. Appl Biochem Biotech 2011;164:1150–9. [DOI] [PubMed] [Google Scholar]
- Liu H, Xu L, Yan M et al. . gTME for construction of recombinant yeast co-fermenting xylose and glucose. Chinese J Biotechnol 2008;24:1010–5. [DOI] [PubMed] [Google Scholar]
- Liu H, Yan M, Lai C et al. . gTME for improved xylose fermentation of Saccharomyces cerevisiae. Appl Biochem Biotech 2010;160:574–82. [DOI] [PubMed] [Google Scholar]
- Liu ZL. Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biot 2011;90:809–25. [DOI] [PubMed] [Google Scholar]
- Liu ZL, Slininger PJ, Dien BS et al. . Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J Ind Microbiol Biotechnol 2004;31:345–52. [DOI] [PubMed] [Google Scholar]
- Lohmeier-Vogel EM, Sopher CR, Lee H. Intracellular acidification as a mechanism for the inhibition by acid hydrolysis-derived inhibitors of xylose fermentation by yeasts. J Ind Microbiol Biotechnol 1998;20:75–81. [Google Scholar]
- McNeil M, Darvill AG, Fry SC et al. . Structure and function of the primary cell walls of plants. Ann Rev Biochem 1984;53:625–63. [DOI] [PubMed] [Google Scholar]
- Maiorella BL, Blanch HW, Wilke CR. Feed component inhibition in ethanolic fermentation by Saccharomyces cerevisiae. Biotechnol Bioeng 1984;26:1155–66. [DOI] [PubMed] [Google Scholar]
- Makanae K, Kintaka R, Makino T et al. . Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res 2013;23:300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M et al. . CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 2015;15:DOI: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansure JJ, Panek AD, Crowe LM et al. . Trehalose inhibits ethanol effects on intact yeast cells and liposomes. Biochim BiophysActa 1994;119:309–16. [DOI] [PubMed] [Google Scholar]
- Margaritis A, Bajpaj P. Effect of sugar concentration in Jerusalem artichoke extract on Kluyveromyces marxianus growth and ethanol production. Appl Environ Microb 1983;45:723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Munoz GA, Kane P. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem 2008;283:20309–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell P, Stratford M, Steels H et al. . Physiological characterization of spoilage strains of Zygosaccharomyces bailii and Zygosaccharomyces rouxii isolated from high sugar environments. Int J Food Microbiol 2007;114:234–42. [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Branduardi P, Dato L et al. . Recombinant protein production in yeasts. Methods Mol Biol 2012;824:329–58. [DOI] [PubMed] [Google Scholar]
- Mayer AF, Hellmuth K, Schlieker H et al. . An expression system matures—a highly efficient and cost-effective process for phytase production by recombinant strains of Hansenula polymorpha. Biotechnol Bioeng 1998;63:373–81. [DOI] [PubMed] [Google Scholar]
- Meijnen JP, Randazzo P, Foulquie-Moreno MR et al. . Polygenic analysis and targeted improvement of the complex trait of high acetic acid tolerance in the yeast Saccharomyces cerevisiae. Biotechnol Biofuels 2016;9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millati R, Niklasson C, Taherzadeh J. Effect of pH, time and temperature of overliming on detoxification of dilute-acid hydrolyzates for fermentation by Saccharomyces cerevisiae. Process Biochem 2002;38:515–22. [Google Scholar]
- Mira NP, Becker JD, Sa-Correia I. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 2010;14:587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira NP, Lourenco AB, Fernandes AR et al. . The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res 2009;9:202–16. [DOI] [PubMed] [Google Scholar]
- Mira NP, Palma M, Guerreiro JF et al. . Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microbial Cell Fact 2010;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira NP, Teixeira MC, Sa-Correia I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 2010;14:525–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modig T, Liden G, Taherzadeh J. Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 2002;363:769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol 2007;27:6446–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraitis C, Curran BP. Reactive oxygen species may influence the heat shock response and stress tolerance in the yeast Saccharomyces cerevisiae. Yeast 2004;21:313–23. [DOI] [PubMed] [Google Scholar]
- Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 2012;190:1157–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai N, Masaki K, Fujii T et al. . PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J Biosci Bioeng 2010;109:564–9. [DOI] [PubMed] [Google Scholar]
- Muzzey D, Gomez-Uribe CA, Mettetal JT et al. . A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell 2009;138:160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendranath NV, Thomas KC, Ingledew WM. Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in minimal medium. J Ind Microbiol Biot 2001;26:171–7. [DOI] [PubMed] [Google Scholar]
- Neves M-J, François J. On the mechanism by which a heat shock induces trehalose accumulation in Saccharomyces cerevisiae. Biochem J 1992;288:859–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol R 2008;72:379–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CY, Jung M, Lee J et al. . Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microbial Cell Fact 2012;11:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng 2010;12:307–31. [DOI] [PubMed] [Google Scholar]
- Nissen TL, Anderlund M, Nielsen J et al. . Expression of a cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast 2001;18:19–32. [DOI] [PubMed] [Google Scholar]
- Nygård Y, Mojzita D, Toivari M et al. . The diverse role of Pdr12 in resistance to weak organic acids. Yeast 2014;31:219–31. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell 2004;15:532–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Nitta A, Uchiyama H et al. . Tolerance mechanism of the ethanoltolerant mutant of sake yeast. J Biosci Bioeng 2000;90:313–20. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Tatsumi H, Murakami S et al. . Secretion of Aspergillus oryzae alkaline protease in an osmophilic yeast, Zygosaccharomyces rouxii. Agr Biol Chem 1990;54:2521–9. [PubMed] [Google Scholar]
- ŌNishi H. Osmophilic Yeasts. Adv Food Res 1963;12:53–94. [PubMed] [Google Scholar]
- Pais TM, Foulquie-Moreno MR, Hubmann G et al. . Comparative polygenic analysis of maximal ethanol accumulation capacity and tolerance to high ethanol levels of cell proliferation in yeast. PLoS Genet 2013;9:e1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Dias PJ, Roque FC et al. . The Zygosaccharomyces bailii transcription factor Haa1 is required for acetic acid and copper stress responses suggesting subfunctionalization of the ancestral bifunctional protein Haa1/Cup2. BMC Genomics 2017;18:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist E, Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresource Technol 2000;74:25–33. [Google Scholar]
- Palmqvist E, Grage H, Meinander NQ et al. . Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol Bioeng 1999;63: 46–55. [DOI] [PubMed] [Google Scholar]
- Pampulha ME, Loureiro-Dias MC. Activity of glycolytic enzymes of Saccharomyces cerevisiae in the presence of acetic acid. Appl Microbiol Biot 1990;34:375–80. [Google Scholar]
- Petersson A, Almeida JR, Modig T et al. . A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 2006;23:455–64. [DOI] [PubMed] [Google Scholar]
- Petitjean M, Teste MA, Francois JM et al. . Yeast tolerance to various stresses relies on the trehalose-6P synthase (Tps1) protein, not on trehalose. J Biol Chem 2015;290:16177–90. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Petitjean M, Teste MA, Leger-Silvestre I et al. . A new function for the yeast trehalose-6P synthase (Tps1) protein, as key pro-survival factor during growth, chronological ageing, and apoptotic stress. Mech Ageing Dev 2016;161:234–46. [DOI] [PubMed] [Google Scholar]
- Pinel D, D’Aoust F, del Cardayre SB et al. . Saccharomyces cerevisiae genome shuffling through recursive population mating leads to improved tolerance to spent sulfite liquor. Appl Environ Microb 2011;77:4736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA et al. . The transcriptional response of yeast to saline stress. J Biol Chem 2000;275:17249–55. [DOI] [PubMed] [Google Scholar]
- Prista C, Soeiro A, Vesely P et al. . Genes from Debaryomyces hansenii increase salt tolerance in Saccharomyces cerevisiae W303. FEMS Yeast Res 2002;2:151–7. [DOI] [PubMed] [Google Scholar]
- Radecka D, Mukherjee V, Mateo RQ et al. . Looking beyond Saccharomyces: the potential of non-conventional yeast species for desirable traits in bioethanol fermentation. FEMS Yeast Res 2015;15:DOI: 10.1093/femsyr/fov053. [DOI] [PubMed] [Google Scholar]
- Ranatunga TD, Jervis J, Helm RF et al. . The effect of overliming on the toxicity of dilute acid pretreated lignocellulosics- the role of inorganics, uronic acids and ether-soluble organics. Enzyme Microb Technol 2000;27:240–7. [DOI] [PubMed] [Google Scholar]
- Rasmussen H, Sorensen HR, Meyer AS. Formation of degradation compounds from lignocellulosic biomass in the biorefinery: sugar reaction mechanisms. Carbohydr Res 2014;385:45–57. [DOI] [PubMed] [Google Scholar]
- Reinders A, Romano I, Wiemken A et al. . The thermophilic yeast Hansenula polymorpha does not require trehalose synthesis for growth at high temperatures but does for normal acquisition of thermotolerance. J Bacteriol 1999;181:4665–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remize F, Roustan JL, Sablayrolles JM et al. . Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl Environ Microb 1999;65:143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JSM, Silva JT, Panek AD. Trehalose metabolism in Saccharomyces cerevisiae during heat-shock. Biochim Biophys Acta 1994;1200:139–47. [DOI] [PubMed] [Google Scholar]
- Rizzi M, Harwart K, Erlemann P et al. . Purification and properties of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng 1989;67:20–4. [Google Scholar]
- Roze LV, Chanda A, Linz JE. Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet Biol 2011;48:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ et al. . Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife 2014;3:e03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales K, Brandt W, Rumbak E et al. . The LEA-like protein HSP 12 in Saccharomyces cerevisiae has a plasma membrane location and protects membranes against desiccation and ethanol-induced stress. Biochim Biophys Acta 2000;1463:267–78. [DOI] [PubMed] [Google Scholar]
- Sanchez OJ, Cardona CA. Trends in biotechnological production of fuel ethanol from different feedstocks. Bioresource Technol 2008;99:5270–95. [DOI] [PubMed] [Google Scholar]
- Sanchez RG, Karhumaa K, Fonseca C et al. . Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol Biofuels 2010;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA et al. . Hsp104 is required for tolerance to many forms of stress. EMBO J 1992;11:1357–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardonini CA, DiBiasio D. A model for growth of Saccharomyces cerevisiae containing a recombinant plasmid in selective media. Biotechnol Bioeng 1987;29:469–75. [DOI] [PubMed] [Google Scholar]
- Sauer M, Branduardi P, Valli M et al. . Production of L-ascorbic acid by metabolically engineered Saccharomyces cerevisiae and Zygosaccharomyces bailii. Appl Environ Microb 2004a;70:6086–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U. Evolutionary engineering of industrially important microbial phenotypes. In: Nielsen JEggeling L, Dynesen J et al. (eds). Metabolic Engineering. Berlin, Heidelberg: Springer; 2001, 29–69. [DOI] [PubMed] [Google Scholar]
- Sauer U, Canonaco F, Heri S et al. . The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 2004b;279:6613–9. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol 2010;61:263–89. [DOI] [PubMed] [Google Scholar]
- Schoondermark-Stolk SA, ter Schure EG, Verrips CT et al. . Identification of salt-induced genes of Zygosaccharomyces rouxii by using Saccharomyces cerevisiae GeneFilters. FEMS Yeast Res 2002;2:525–32. [DOI] [PubMed] [Google Scholar]
- Schwan RF, Cooper RM, Wheals AE. Endopolygalacturonase secretion by Kluyveromyces marxianus and other cocoa pulp-degrading yeasts. Enzyme Microb Technol 1997;21:234–44. [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM et al. . Interdependence of cell growth and gene expression—origins and consequences. Science 2010;330:1099–102. [DOI] [PubMed] [Google Scholar]
- Scott M, Hwa T. Bacterial growth laws and their applications. Curr Opin Biotechnol 2011;22:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Klumpp S, Mateescu EM et al. . Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol 2014;10:747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre AV, Murray AW, Leu JY. High-resolution mutation mapping reveals parallel experimental evolution in yeast. PLoS Biol 2006;4:e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahsavarani H, Sugiyama M, Kaneko Y et al. . Superior thermotolerance of Saccharomyces cerevisiae for efficient bioethanol fermentation can be achieved by overexpression of RSP5 ubiquitin ligase. Biotechnol Adv 2012;30:1289–300. [DOI] [PubMed] [Google Scholar]
- Shapouri H, Gallagher P. USDA’s 2002 Ethanol Cost-of-Production Survey. Agriculture Economic Report Number 841.United States Department of Agriculture; 2005. [Google Scholar]
- Sheng J, Feng X. Metabolic engineering of yeast to produce fatty acid-derived biofuels: bottlenecks and solutions. Front Microbiol 2015;6:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DJ, Wang CL, Wang KM. Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 2009;36:139–47. [DOI] [PubMed] [Google Scholar]
- Shi S, Liang Y, Zhang MM et al. . A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab Eng 2016;33:19–27. [DOI] [PubMed] [Google Scholar]
- Siddiqui MS, Thodey K, Trenchard I et al. . Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res 2012;12:144–70. [DOI] [PubMed] [Google Scholar]
- Silverstein RA, Chen Y, Sharma-Shivappa RR et al. . A comparison of chemical pretreatment methods for improving saccharification of cotton stalks. Bioresource Technol 2007;98:3000–11. [DOI] [PubMed] [Google Scholar]
- Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell 1998;1:639–48. [DOI] [PubMed] [Google Scholar]
- Singleton C, Howard TP, Smirnoff N. Synthetic metabolons for metabolic engineering. J Exp Bot 2014;65:1947–54. [DOI] [PubMed] [Google Scholar]
- Sonderegger M, Sauer U. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microb 2003;69:1990–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere PA. Complexes of sequential metabolic enzymes. Ann Rev Biochem 1987;56: 89–124. [DOI] [PubMed] [Google Scholar]
- Srienc F, Campbell JL, Bailey JE. Analysis of unstable recombinant Saccharomyces cerevisiae population growth in selective medium. Biotechnol Bioeng 1986;28:996–1006. [DOI] [PubMed] [Google Scholar]
- Stampe E, Alcock R, Westby C et al. . Energy consumption of a farm-scale ethanol distillation system. Energ Agr 1983;2:355–68. [Google Scholar]
- Stanley D, Bandara A, Fraser S et al. . The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol 2010a;109:13–24. [DOI] [PubMed] [Google Scholar]
- Stanley D, Fraser S, Chambers PJ et al. . Generation and characterisation of stable ethanol-tolerant mutants of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 2010b;37:139–49. [DOI] [PubMed] [Google Scholar]
- Steensels J, Snoek T, Meersman E et al. . Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol Rev 2014;38:947–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science 2007;315:801–4. [DOI] [PubMed] [Google Scholar]
- Stephen JD, Mabee WE, Saddler JN. Will second-generation ethanol be able to compete with first-generation ethanol? Opportunities for cost reduction. Biofue Bioprod Bior 2012; 6:159–76. [Google Scholar]
- Stovicek V, Borodina I, Forster J. CRISPR–Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab Eng Commun 2015;2:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Matsuda F, Hasunuma T et al. . Implementation of a transhydrogenase-like shunt to counter redox imbalance during xylose fermentation in Saccharomyces cerevisiae. Appl Microbiol Biot 2013;97:1669–78. [DOI] [PubMed] [Google Scholar]
- Swinnen S, Schaerlaekens K, Pais T et al. . Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res 2012;22:975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen S, Thevelein JM, Nevoigt E. Genetic mapping of quantitative phenotypic traits in Saccharomyces cerevisiae. FEMS Yeast Res 2012;12:215–27. [DOI] [PubMed] [Google Scholar]
- Taherzadeh MJ, Gustafsson L, Niklasson C et al. . Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl Microbiol Biot 2000;53:701–8. [DOI] [PubMed] [Google Scholar]
- Takagi H, Takaoka M, Kawaguchi A et al. . Effect of L-proline on sake brewing and ethanol stress in Saccharomyces cerevisiae. Appl Environ Microb 2005;71:8656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamás MJ, Rep M, Thevelein JM et al. . Stimulation of the yeast high osmolarity glycerol (HOG) pathway-evidence for a signal generated by a change in turgor rather than bywater stress. FEBS Lett 2000;472:159–65. [DOI] [PubMed] [Google Scholar]
- Teunissen A, Dumortier F, Gorwa MF et al. . Isolation and characterization of a freeze-tolerant diploid derivative of an industrial Baker's yeast strain and its use in frozen doughs. Appl Environ Microb 2002;68:4780–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas-Pejo E, Ballesteros M, Oliva JM et al. . Adaptation of the xylose fermenting yeast Saccharomyces cerevisiae F12 for improving ethanol production in different fed-batch SSF processes. J Ind Microbiol Biotechnol 2010;37:1211–20. [DOI] [PubMed] [Google Scholar]
- Trantas E, Panopoulos N, Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng 2009;11:355–66. [DOI] [PubMed] [Google Scholar]
- Tsai CS, Kong II, Lesmana A et al. . Rapid and marker-free refactoring of xylose-fermenting yeast strains with Cas9/CRISPR. Biotechnol Bioeng 2015;112:2406–11. [DOI] [PubMed] [Google Scholar]
- Tsuge Y, Kawaguchi H, Sasaki K et al. . Engineering cell factories for producing building block chemicals for bio-polymer synthesis. Microb Cell Fact 2016;15:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah A, Orij R, Brul S et al. . Quantitative analysis of the modes of growth inhibition by weak organic acids in Saccharomyces cerevisiae. Appl Environ Microb 2012;78:8377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maris AJA, Winkler AA, Kuyper M et al. . Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key Component. Adv Biochem Eng Biotechnol 2007;108:179–204. [DOI] [PubMed] [Google Scholar]
- van Rensburg E, den Haan R, Smith J et al. . The metabolic burden of cellulase expression by recombinant Saccharomyces cerevisiae Y294 in aerobic batch culture. Appl Microbiol Biot 2012;96:197–209. [DOI] [PubMed] [Google Scholar]
- van Voorst F, Houghton-Larsen J, Jonson L et al. . Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 2006;23:351–9. [DOI] [PubMed] [Google Scholar]
- Veine DM, Arscott LD, Willians Jr. CH. Redox potentials for yeast, Escherichia coli and human glutathione reductase relative to the NAD+/NADH redox couple- enzyme forms active in catalysis. Biochemistry 1998;37:15575–82. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Van Kleef R, Frank J et al. . Properties of the NAD(P)H-dependent xylose reductase from the xylosefermenting yeast Pichia stipitis. Biochem J 1985;226:669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y et al. . Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol R 2012;76:115–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas JV, Petridis L, Qi X et al. . Mechanism of lignin inhibition of enzymatic biomass deconstruction. Biotechnol Biofuels 2015;8:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vind J, Sørensen MA, Rasmussen MD et al. . Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes. J Mol Biol 1993;231:678–88. [DOI] [PubMed] [Google Scholar]
- Walker GM, Maynard A. Accumulation of magnesium ions during fermentative metabolism in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 1997;18:1–3. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Abu Saleh A, Pack SP et al. . Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 2007a;153:3044–54. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Saleh AA, Pack SP et al. . Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+-dependent xylitol dehydrogenase. J Biotechnol 2007b;130:316–9. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Tamura K, Magbanua JP et al. . Elevated expression of genes under the control of stress response element (STRE) and Msn2p in an ethanol-tolerance sake yeast Kyokai no.11 J Biosci Bioeng 2007c;104:163–70. [DOI] [PubMed] [Google Scholar]
- Wei N, Quarterman J, Jin YS. Marine macroalgae: an untapped resource for producing fuels and chemicals. Trends Biotechnol 2013;31:70–77. [DOI] [PubMed] [Google Scholar]
- Welch J, Fogel S, Buchman C et al. . The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. EMBO J 1989;8:255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker S, Rudolph B, Frenzel E et al. . Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol Cell 2010;39:507–20. [DOI] [PubMed] [Google Scholar]
- Wheals AE, Basso LC, Alves DMG et al. . Fuel ethanol after 25 years. Trends Biotechnol 1999;17:482–7. [DOI] [PubMed] [Google Scholar]
- Wi SG, Kim HJ, Mahadevan SA et al. . The potential value of the seaweed Ceylon moss (Gelidium amansii) as an alternative bioenergy resource. Bioresource Technol 2009;100:6658–60. [DOI] [PubMed] [Google Scholar]
- Wiemken A. Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie Leeuw 1990;58:209–17. [DOI] [PubMed] [Google Scholar]
- Wisselink HW, Toirkens MJ, del Rosario Franco Berriel M et al. . Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of L-arabinose. Appl Environ Microb 2007;73:4881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JM, Yang KM, Kim SU et al. . High temperature stimulates acetic acid accumulation and enhances the growth inhibition and ethanol production by Saccharomyces cerevisiae under fermenting conditions. Appl Microbiol Biot 2014;98:6085–94. [DOI] [PubMed] [Google Scholar]
- Wright J, Bellissimi E, de Hulster E et al. . Batch and continuous culture-based selection strategies for acetic acid tolerance in xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res 2011;11:299–306. [DOI] [PubMed] [Google Scholar]
- Wythes JR, Wainwright DH, Blight GW. Nutrient composition of Queensland molasses. Aust J Exp Agr Anim Husbandry 1978;18:629–34. [Google Scholar]
- Yang J, Bae JY, Lee YM et al. . Construction of Saccharomyces cerevisiae strains with enhanced ethanol tolerance by mutagenesis of the TATA-binding protein gene and identification of novel genes associated with ethanol tolerance. Biotechnol Bioeng 2011;108:1776–87. [DOI] [PubMed] [Google Scholar]
- Yang Y, Foulquie-Moreno MR, Clement L et al. . QTL analysis of high thermotolerance with superior and downgraded parental yeast strains reveals new minor QTLs and converges on novel causative alleles involved in RNA processing. PLoS Genet 2013;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KM, Rosenfield CL, Knipple DC. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl Environ Microb 2003;69:1499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AQ, Juwono NK, Foo JL et al. . Metabolic engineering of Saccharomyces cerevisiae for the overproduction of short branched-chain fatty acids. Metab Eng 2016;34:36–43. [DOI] [PubMed] [Google Scholar]
- Zacchi G, Axelsson A. Economic evaluation of preconcentration in production of ethanol from dilute sugar solutions. Biotechnol Bioeng 1989;34:223–33. [DOI] [PubMed] [Google Scholar]
- Zha Y, Muilwijk B, Coulier L. Inhibitory compounds in lignocellulosic biomass hydrolysates during hydrolysate fermentation processes. J Bioprocess Biotechniq 2012;2:1000112. [Google Scholar]
- Zhang W, Geng A. Improved ethanol production by a xylosefermenting recombinant yeast strain constructed through a modified genome shuffling method. Biotechnol Biofuels 2012;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li S, Li L et al. . Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J Am Chen Soc 2006;128:13030–1. [DOI] [PubMed] [Google Scholar]
- Zheng DQ, Wu XC, Tao XL et al. . Screening and construction of Saccharomyces cerevisiae strains with improved multi-tolerance and bioethanol fermentation performance. Bioresource Technol 2011a;102:3020–7. [DOI] [PubMed] [Google Scholar]
- Zheng DQ, Wu XC, Wang PM et al. . Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 2011b;38:415–22. [DOI] [PubMed] [Google Scholar]
- Zhou H, Cheng JS, Wang BL et al. . Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 2012;14:611–22. [DOI] [PubMed] [Google Scholar]