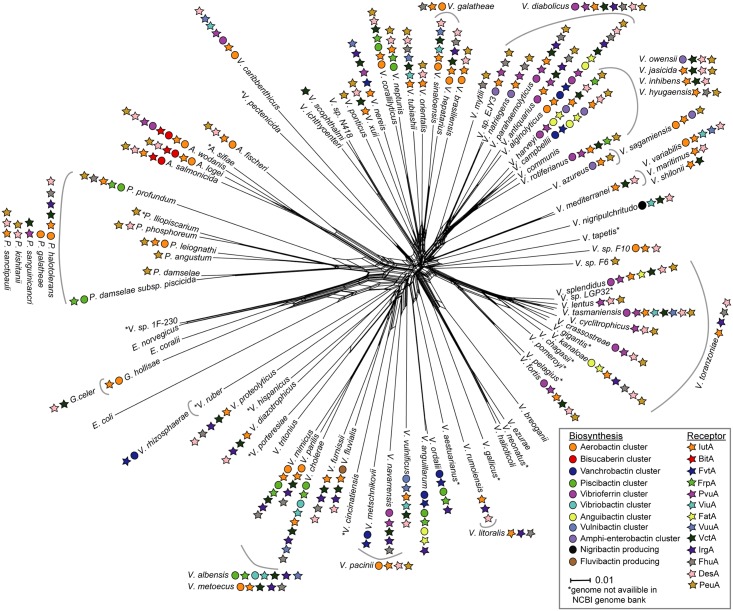
Fig 2. Distribution of homologs of known Vibrionaceae siderophore biosynthesis clusters and receptors mapped to a phylogeny.
The phylogenetic split network is based on a dataset from Sawabe and co-workers [8], and consists of the genes ftsZ, gap, gyrB, mreB, pyrH, recA, rpoA and topA. The tree was constructed using SplitsTree4 to concatenate the individual gene alignments, and settings for network were uncorrected P and NeighborNet [56]. Branch lengths are to scale and species located outside grey arches were not included in the MLSA files and have been placed according to literature [71–86].