Fig 3. Phylogeny of the piscibactin biosynthesis cluster and receptor within the Vibrionaceae family.
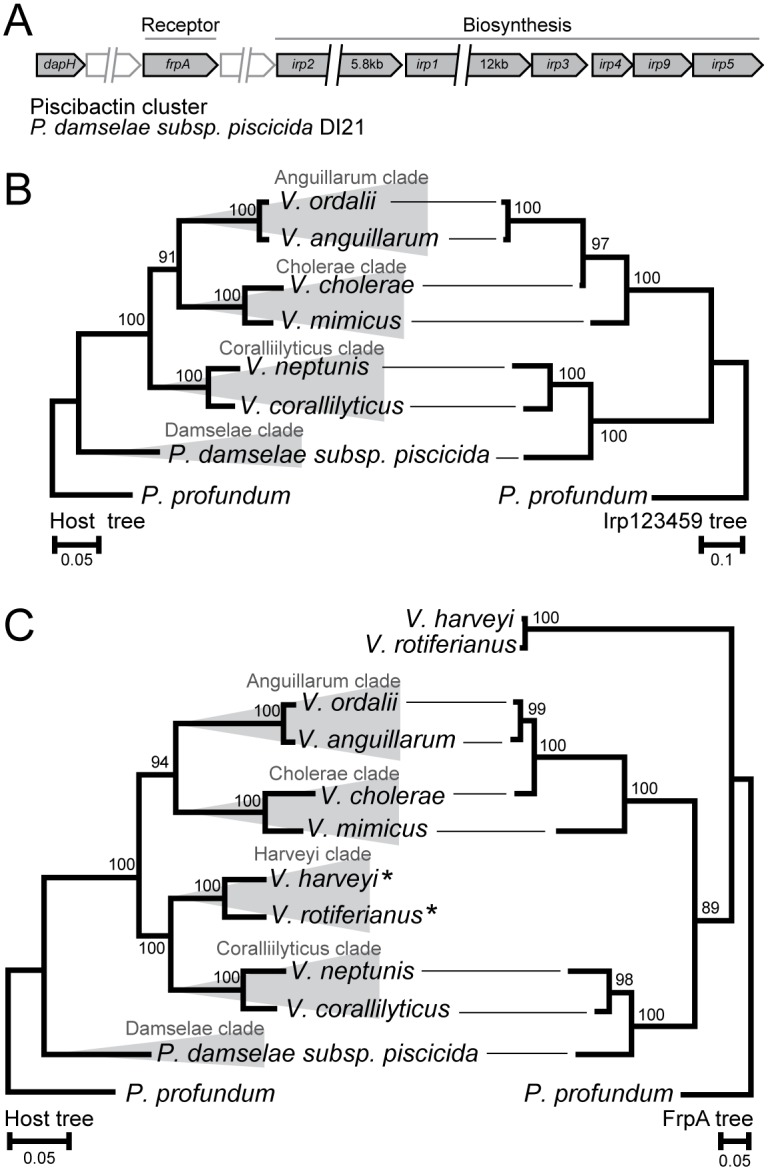
(A) The cluster organization of the biosynthesis cluster and the cognate receptor. (B) Host phylogeny on the left and piscibactin biosynthesis system (Irp123459) phylogeny on the right. (C) Host phylogeny on the left and piscibactin receptor (FrpA) phylogeny on the right. Asterisks denote species that do not encode the piscibactin biosynthesis system, i.e., the FrpA homolog is an exogenous siderophore receptor. Evolutionary analyses were conducted in MEGA6 [58]. The host trees were generated using the ML method and the TM model [59]. The siderophore biosynthesis cluster and receptor trees were generated using the ML method and the JTT model [87]. Bootstrap values are shown at the nodes (JTT model, 2000 replicates) [88]. Branch lengths are measured substitutions per site.
