Fig. 1. Evolution of phage T4 genome under CRISPR-Cas9 pressure.
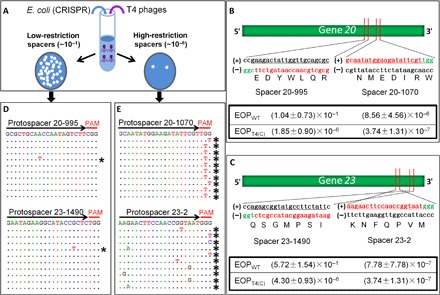
(A) Experimental scheme for testing the effect of CRISPR-Cas on phage T4 infection. Efficient cleavage by Cas9 nuclease at the protospacer sequence disrupts the phage genome, resulting in loss of plaque-forming ability (plate on the right; high-restriction spacer). Inefficient cleavage by Cas9 nuclease reduces plating efficiency (plate on the left; low-restriction spacer). (B and C) Plating efficiencies of high-restriction spacers 20-1070 and 23-2 and low-restriction spacers 20-995 and 23-1490. Shown are the locations of spacers on genes 20 and 23 and the nucleotide and amino acid sequences corresponding to the protospacer (red) and PAM (green) sequences. The sequences of the complementary strand are shown in black. Efficiency of plating (EOP) was determined, as described in Materials and Methods. The data shown are the average of three independent experiments ± SD. (D and E) Alignment of sequences corresponding to single plaques produced from infection of various spacer-expressing E. coli. The DNA from single plaques was amplified and sequenced, as described in Materials and Methods. The black arrows correspond to the spacer sequences, and the red lines correspond to the PAM sequences. The sequence at the top of each panel corresponds to the WT sequence. The dotted lines below correspond to the sequence obtained from each plaque. Only the mutated nucleotides are shown. The asterisks indicate the mutant sequences.
