Highlights
-
•
Routine strategies for hepatitis C virus (HCV) genotyping have several limitations. Deep sequencing methods can solve this problem.
-
•
Accurate determination of viral genotypes and subtypes would allow optimal patient management and the most effective therapy.
-
•
Mixed infections may represent a key factor for efficient therapy. Patients infected with more than one HCV genotype (mixed infection) can be detected only by deep sequencing methods.
-
•
These patients can fail treatment with direct-acting antiviral agents, hence next-generation sequencing methods are highly recommended in clinical practice.
Keywords: HCV, Mixed infection, Deep sequencing, NGS, Direct-acting antivirals
Abstract
Background
The effectiveness of the new generation of hepatitis C treatments named direct-acting antiviral agents (DAAs) depends on the genotype, subtype, and resistance-associated substitutions present in individual patients. The aim of this study was to evaluate a massive sequencing platform for the analysis of genotypes and subtypes of hepatitis C virus (HCV) in order to optimize therapy.
Methods
A total of 84 patients with hepatitis C were analyzed. The routine genotyping methodology for HCV used at the study institution (Versant HCV Assay, LiPA) was compared with a deep sequencing platform (454/GS-Junior and Illumina MiSeq).
Results
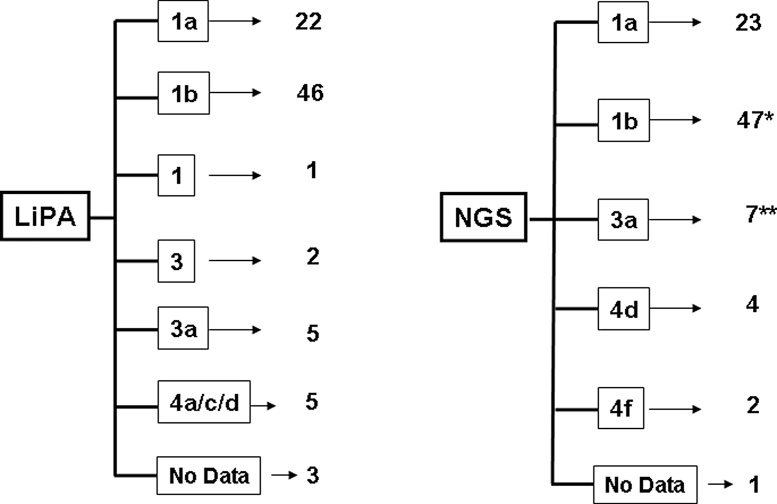
The mean viral load in these HCV patients was 6.89 × 106 ± 7.02 × 105. Viral genotypes analyzed by LiPA were distributed as follows: 26% genotype 1a (22/84), 55% genotype 1b (46/84), 1% genotype 1 (1/84), 2.5% genotype 3 (2/84), 6% genotype 3a (5/84), 6% genotype 4a/c/d (5/84). When analyzed by deep sequencing, the samples were distributed as follows: 27% genotype 1a (23/84), 56% genotype 1b (47/84), 8% genotype 3a (7/84), 5% genotype 4d (4/84), 2.5% genotype 4f (2/84). Six of the 84 patients (7%) were infected with more than one subtype. Among these, 33% (2/6) failed DAA-based triple therapy.
Conclusions
The detection of mixed infection could explain some treatment failures. Accurate determination of viral genotypes and subtypes would allow optimal patient management and improve the effectiveness of DAA therapy.
Introduction
An estimated 170 million individuals worldwide are chronically infected with hepatitis C virus (HCV), and according to the US Centers for Disease Control and Prevention, approximately 10% to 20% of them will develop severe liver disease (https://wwwnc.cdc.gov/travel/yellowbook/2018/infectious-diseases-related-to-travel/hepatitis-c). The complex genetic variability of HCV has been classified into four hierarchical strata: genotype, subtype, isolate, and quasispecies (Farci and Purcell, 2000). A consensus proposal for HCV classification was initially presented in 2005 (Simmonds et al., 2005) and updated in 2014 (Smith et al., 2014). To date, seven HCV genotypes have been confirmed, with whole-genome nucleotide sequences differing by >30%, and each can be further subdivided into related subtypes (67 confirmed), with nucleotide sequence divergence of between 15% and 30% (Smith et al., 2014).
Genotype identification has long been used in clinical practice. The different genotypes and subtypes of HCV show different responses to antiviral therapy with both the traditional treatment of pegylated interferon α/ribavirin and the currently available HCV direct-acting antiviral (DAA) drugs (Legrand-Abravanel et al., 2009, Poveda et al., 2014). Therefore, accurate genotyping and subtyping of HCV is needed for the selection of the appropriate treatment regimen. The quasispecies provide a large pool of genetic variants that can adapt to new selection pressures such as the host’s cell defence and antiviral treatments, resulting in chronic infection and HCV drug resistance (Tarr et al., 2015). However, subtype identification has been used mainly in epidemiological studies. Both in vitro studies and clinical trials with different classes of DAA (NS3 protease inhibitors, NS5A inhibitors, and nucleos(t)ide and non-nucleos(t)ide NS5B polymerase inhibitors), given with pegylated interferon α/ribavirin or in interferon-free combinations, have shown lower response rates for HCV genotype 1a than for HCV genotype 1b (Poordad et al., 2014).
Currently available genotyping methods based on reverse hybridization with subtype-specific primers and probes targeting the 5′ untranscribed region (UTR) and core regions (Versant HCV Genotype 2.0 system; Siemens), as well as real-time PCR assays based on the 5′ UTR and NS5B sequencing (Abbott HCV Genotype II assay), accurately differentiate major HCV genotypes in the majority of cases and are widely used because of their technical simplicity. However, these assays have not been designed to confidently identify mixed infections, and their ability to accurately discriminate HCV subtypes other than 1a and 1b is very limited (Chevaliez et al., 2009, Avó et al., 2013, González et al., 2013).
A subtyping assay based on phylogenetic analysis of reads obtained by deep sequencing of an NS5B fragment has recently been developed (Quer et al., 2015). The aim of this study was to evaluate the effectiveness of this massive sequencing methodology for the accurate classification of HCV in the real-life setting, in order to optimize antiviral therapy.
Methods
All patients participating in this study signed an informed consent agreement. Serum samples from 84 HCV-infected patients were used, independent of clinical information. All specimens were collected and processed as recommended and stored at −80 °C. The study was approved by the Ethics Committee of Valme University Hospital. This study was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines.
HCV RNA was extracted from 650 μl of plasma or serum by automated RNA extraction using a total nucleic acid isolation (TNAI) kit (AmpliPrep System; Roche Diagnostics, Barcelona, Spain), or by manual RNA extraction (150 μl) using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany).
Routine HCV genotyping was performed using the HCV Genotype 2.0 assay (LiPA HCV v.2.0; Siemens Healthcare Diagnostics, Eragny, France). This system is a line probe assay based on reverse hybridization of biotinylated products targeting the 5′ UTR and a fragment of the core region, which are hybridized to immobilized oligonucleotide probes in nitrocellulose strips that are specific for the 5′-UTR core regions of the six genotypes.
The NS5B target for HCV subtyping was a 339-nucleotide (nt) region spanning nt 8280 to nt 8618, according to data for isolate H77 available in GenBank (accession number AF009606). Primer and PCR condition data have been deposited under patent number EP13382278. Further details of the PCR methods (primers sequences, etc.) have been reported previously by Quer et al. (2015).
Deep sequencing
After nested PCR amplification, the final product is flanked by universal M13 sequences at both ends. The last PCR amplification is performed in a 96-well plate previously preloaded with a lyophilized universal primer pair consisting of an M13 universal primer plus MID plus oligonucleotide A or B (454 sequencing) (M13-MID_A/B primer; TIB Molbiol, Berlin, Germany). Each patient sample requires a different MID sequence. The PCR product of this amplification is then automatically purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA) and quantified using a fluorometric titration Quant-iT PicoGreen double-stranded DNA (dsDNA) kit (Life Technologies, Eugene, OR, USA) in an Infinity fluorometer (Tecan, Männedorf, Switzerland), with equimolecular pooling and a final enrichment step using an automated REMe STARlet Hamilton workstation (Hamilton Robotics, Reno, NV, USA).
Massive sequencing was performed using the 454/GS-Junior platform (Roche, Branford, CT, USA) and titanium chemistry (GS-Junior Titanium Sequencing kit), which enables sequencing of 400–500-nt fragments. An average of 1250 sequences (reads) was obtained (minimum 200, maximum 5240).
The FASTA file from the GS-Junior was demultiplexed to obtain a FASTA file for each sample and strand. Sequences not covering the full amplicon (positions 8280 to 8618) or showing more than 2 Ns or 3 gaps were discarded. The threshold of 90% identity was selected on the basis of the minimum observed distance between pairs of reference sequences of different subtypes.
Statistical analysis
Rates and proportions were analyzed by binomial test and Fisher’s exact test. A p-value of <0.05 was considered statistically significant.
Results
Patients attending the outpatient clinic of Valme University Hospital (January 2015–May 2016) with a chronic active HCV infection were selected and classified according to the HCV genotype determined using the HCV Genotype 2.0 assay (LiPA HCV v.2.0). All patient data were coded to maintain anonymity. Serum samples obtained from patients before treatment were used to determine viral load and genotypes. The mean viral load was 6.89 × 106 ± 7.02 × 105.
A total of 84 patients were included; 51 were male and 33 were female. Their average age at baseline was 53.8 ± 8.3 years. Thirty-five patients (42%) received triple therapy for HCV (pegylated interferon + ribavirin + telaprevir/boceprevir). Forty-nine patients (58%) received sofosbuvir-based therapy. Seventeen (14%) patients did not respond to antiviral therapy: 15 did not respond to triple therapy and two did not respond to sofosbuvir-based therapy.
The LiPA assay failed (no data after analysis) for three of the 84 samples tested (3.5%), while the ultra-deep sequencing system failed for one sample (1%), due to degraded RNA. Table 1 shows the major differences found in the identification of genotypes on comparison of the routine method with the deep sequencing technology: the HCV genotype was not properly identified for eight patients (10%) when using the routine method. The 454 technology has recently been discontinued by Roche. Therefore, the samples of a subset of 24 patients were re-analyzed: the same analysis was performed with the Illumina MiSeq platform. The same results (in terms of high-resolution genotyping) were found for this group of patients (n = 24) when compared to the data obtained with the 454/GS-Junior platform.
Table 1.
Comparison of results obtained with the routine method (patients with indeterminate genotypes) and deep sequencing genotyping.
| Patient code | Viral load | Versant HCV genotyping | Deep sequencing subtyping (GS-Junior) |
|---|---|---|---|
| 327-20 | 1 430 000 | 4* | 4d |
| 327-25 | 246 000 | 4* | 4d |
| 68648 | 562 000 | 3* | 3a |
| 364956 | 6 040 000 | 1a | 1b** |
| 7217 | 548 000 | 1* | 1a + 1b |
| 327-61 | 1 620 000 | 4* | 4d |
| 72-69060 | 656 000 | 1a | 1a + 1b |
| 57946 | 59 900 | 1b | 1a** |
HCV, hepatitis C virus.
Indeterminate sub-genotype.
Routine method failed in HCV genotyping.
The HCV genotype distributions for the study patients according to the genotyping method used – routine (LiPA) and deep sequencing – are shown in Figure 1. The main difference was found for patients infected with HCV genotypes 3 and 4, which could not be classified into subtypes using the routine method. For two more samples, the subtype could not be identified using LiPA. This problem was resolved when using the deep sequencing method.
Figure 1.
HCV genotype distribution in patients according to the genotyping method. The LiPA method failed in the genotyping of three of the 84 patients. Deep sequencing failed for one patient. *Mixed infections were detected, with a majority of genotype 1b. **Two mixed infections 1b + 3a. See Table 2 for details.
A typical deep sequencing analysis output file from the analysis of 20 samples is given in the Supplementary Material (Figure S1).
The deep sequencing method was able to identify patients with HCV mixed infections (more than one HCV genotype) (Table 2). Massive sequencing analysis revealed that six of the 84 patients (7%) were infected with more than one subtype simultaneously. Four of them (4/6, 67%) were infected with subtypes 1a and 1b, while the other two (2/6, 33%) were infected with subtypes 1b and 3a.
Table 2.
Mixed infections detected in HCV patients using the deep sequencing method.
| Patient code | Viral load | Genotypes detected |
|---|---|---|
| 7217 | 548 000 | 1a (2%) + 1b (98%)a |
| 72-69060 | 656 000 | 1a (1%) + 1b (99%) |
| 62205 | 3 670 000 | 1a (1%) + 1b (99%) |
| 696561 | 2 900 000 | 1a (1%) + 3a (99%)b |
| 707400 | 583 000 | 1b (99%) + 3a (1%) |
| 331235 | 6 892 395 | 1a (3%) + 1b (97%) |
HCV, hepatitis C virus.
This patient failed treatment with sofosbuvir + ledipasvir.
This patient failed treatment with sofosbuvir + daclatasvir.
Interestingly, two of the six patients with mixed infection (33%) failed DAA-based triple anti-HCV therapy. These two samples were analyzed using population Sanger sequencing and no resistance-associated substitutions for DAA inhibitors were detected (F. García, personal communication). Mixed genotype HCV infections cannot be detected using routine methods. Deep sequencing is the only method available that can identify these.
Discussion
Some DAAs used to treat HCV infections act in a genotype-specific manner (McNaughton et al., 2014, Bagaglio et al., 2016, Patiño-Galindo et al., 2016). The potential outcome of DAA treatment regimes used on mixed HCV infections, i.e. concurrent infections with more than one HCV genotype, has not been considered in the clinical guidelines. This is probably because commercially available genotyping methods are only capable of identifying the dominant genotype present within a mixed infection sample, leaving minor genotypes undetected.
In this study, it was demonstrated that the routine method (LiPA) failed to accurately subtype seven out of 84 infections (8.5%). This is not a minor issue from the clinical view point, since this information is useful for clinical diagnosis, guidance of clinical treatment, patient management, and HCV epidemiology studies (Tong et al., 2015, Nieto-Aponte et al., 2017).
An added benefit of deep sequencing techniques is the possibility of detecting mixed infections. In this study, six mixed infections were detected in 84 samples (7%) (Table 2). Other studies have found mixed HCV infections at rates from 5% to 25.3% (Pham et al., 2010, Thomson et al., 2011). Moreover, DAA treatment in patients with mixed infections may be associated with the occurrence of genotype switching, whereby a previously undetected minority variant drug-resistant genotype expands to replace the successfully treated majority variant genotype (Quer et al., 2016). Further work is required to assess the impact of minority variant strains on patients treated with DAA therapy.
In the present study, most of the patients received triple therapy with boceprevir or telaprevir. Two of the patients who were found to have mixed infections failed therapy with sofosbuvir + ledipasvir and sofosbuvir + daclatasvir (Table 2). Using population Sanger sequencing, no resistance-associated substitutions relevant to DAA antivirals were found. Genotype diversity also directly affects the outcome of infected patients, due to genotype-specific differences in response to treatment and disease severity (EASL, 2015). Hence, several more effective and better tolerated DAAs are now in clinical development (Wendt et al., 2014). Very recently, it was shown that NS5A resistance-associated substitutions in patients with genotype 1a HCV had a negative impact on the treatment outcome (Zeuzem et al., 2017). The present authors are currently developing a deep sequencing protocol to detect HCV resistance mutations in the NS3, NS5A, and NS5B regions.
In conclusion, subtyping analysis based on deep sequencing is much more reliable and useful than the systems currently used in routine clinical practice. However, deep sequencing methods are still expensive compared to routine methods, and a cost-effectiveness analysis is required before the wide implementation of this technique in clinical practice. The accurate determination of viral genotypes and subtypes would allow optimal patient management and the most effective therapy. Moreover, the detection of mixed infections could explain the failure of some therapies.
Ethical approval and consent to participate
This study was approved by the Ethics Committee of Valme University Hospital. This study was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines.
Availability of data and material
All data generated or analysed during this study are included in this published article.
Funding
This work was supported by grants from the Andalusian Government (#PI0892-2012) and Instituto de Salud Carlos III (PI14/01349, PI16/00337) co-financed by the European Regional Development Fund (ERDF).
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by grants from the Andalusian Government (#PI0892-2012) and Instituto de Salud Carlos III (PI14/01349, PI16/00337) co-financed by the European Regional Development Fund (ERDF). J.A. del Campo is supported by the Nicolás Monardes Programme of the Servicio Andaluz de Salud (SAS).
Corresponding Editor: Eskild Petersen, Aarhus, Denmark
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ijid.2017.12.016.
Appendix A. Supplementary data
The following are Supplementary data to this article:
(A) Total number of filtered reads obtained for each sample. (B) UPGMA phylogenetic tree of the NS5B segment of 339 nucleotides from the reference sequences used to classify HCV isolates into the 67 subtypes accepted in 2014 (Smith et al., 2014). Bootstrap confidence levels are included on the branches (blue numbers). Genotype 1 references are in dark green, genotype 2 in dark orange, genotype 3 in blue, genotype 4 in pink, genotype 5 in light green, and genotype 6 in yellow.
References
- Avó A.P., Água-Doce I., Andrade A., Pádua E. Hepatitis C virus subtyping based on sequencing of the C/E1 and NS5B genomic regions in comparison to a commercially available line probe assay. J Med Virol. 2013;85:815–822. doi: 10.1002/jmv.23545. [DOI] [PubMed] [Google Scholar]
- Bagaglio S., Andolina A., Merli M., Uberti-Foppa C., Morsica G. Frequency of natural resistance within NS5a replication complex domain in hepatitis C genotypes 1a, 1b: possible implication of subtype-specific resistance selection in multiple direct acting antivirals drugs combination treatment. Viruses. 2016;8:91. doi: 10.3390/v8040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevaliez S., Bouvier-Alias M., Brillet R., Pawlotsky J.-M. Hepatitis C virus (HCV) genotype 1 subtype identification in new HCV drug development and future clinical practice. Lindenbach B., editor. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Association for Study of Liver EASL recommendations on treatment of hepatitis C 2015. J Hepatol. 2015;63:199–236. doi: 10.1016/j.jhep.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Farci P., Purcell R.H. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin Liver Dis. 2000;20:103–126. [PubMed] [Google Scholar]
- González V., Gomes-Fernandes M., Bascuñana E. Accuracy of a commercially available assay for HCV genotyping and subtyping in the clinical practice. J Clin Virol. 2013;58:249–253. doi: 10.1016/j.jcv.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Legrand-Abravanel F., Colson P., Leguillou-Guillemette H. Influence of the HCV subtype on the virological response to pegylated interferon and ribavirin therapy. J Med Virol. 2009;81:2029–2035. doi: 10.1002/jmv.21583. [DOI] [PubMed] [Google Scholar]
- McNaughton A.L., Thomson E.C., Templeton K., Gunson R.N., Leitch E.C.M. Mixed genotype hepatitis C infections and implications for treatment. Hepatology. 2014;59:1209. doi: 10.1002/hep.26544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Aponte L., Quer J., Ruiz-Ripa A. Assessment of a novel automatic real-time PCR assay on the Cobas 4800 analyzer as a screening platform for hepatitis C virus genotyping in clinical practice: comparison with massive sequencing. J Clin Microbiol. 2017;55:504–509. doi: 10.1128/JCM.01960-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño-Galindo J.Á., Salvatierra K., González-Candelas F., López-Labrador F.X. Comprehensive screening for naturally occurring hepatitis C virus resistance to direct-acting antivirals in the NS3, NS5A, and NS5B genes in worldwide isolates of viral genotypes 1 to 6. Antimicrob Agents Chemother. 2016;60:2402–2416. doi: 10.1128/AAC.02776-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham S.T., Bull R.A., Bennett J.M. Frequent multiple hepatitis C virus infections among injection drug users in a prison setting. Hepatology. 2010;52:1564–1572. doi: 10.1002/hep.23885. [DOI] [PubMed] [Google Scholar]
- Poordad F., Hezode C., Trinh R. ABT-450/r–ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N Engl J Med. 2014;370:1973–1982. doi: 10.1056/NEJMoa1402869. [DOI] [PubMed] [Google Scholar]
- Poveda E., Wyles D.L., Mena Á., Pedreira J.D., Castro-Iglesias Á., Cachay E. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res. 2014;108:181–191. doi: 10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- Quer J., Gregori J., Rodríguez-Frias F. High-resolution hepatitis C virus subtyping using NS5B deep sequencing and phylogeny, an alternative to current methods. Tang Y.-W., editor. J Clin Microbiol. 2015;53:219–226. doi: 10.1128/JCM.02093-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quer J., Rodríguez-Frias F., Gregori J. Deep sequencing in the management of hepatitis virus infections. Virus Res. 2017;239:115–125. doi: 10.1016/j.virusres.2016.12.020. S0168-1702(16)30456-7. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Bukh J., Combet C. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- Smith D.B., Bukh J., Kuiken C. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr A.W., Khera T., Hueging K. Genetic diversity underlying the envelope glycoproteins of hepatitis C Virus: structural and functional consequences and the implications for vaccine design. Viruses. 2015;7:3995–4046. doi: 10.3390/v7072809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E.C., Fleming V.M., Main J. Predicting spontaneous clearance of acute hepatitis C virus in a large cohort of HIV-1-infected men. Gut. 2011;60:837–845. doi: 10.1136/gut.2010.217166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y.-Q., Liu B., Liu H. Accurate genotyping of hepatitis C virus through nucleotide sequencing and identification of new HCV subtypes in China population. Clin Microbiol Infect. 2015;21 doi: 10.1016/j.cmi.2015.05.034. 874.e9-874.e21. [DOI] [PubMed] [Google Scholar]
- Wendt A., Adhoute X., Castellani P. Chronic hepatitis C: future treatment. Clin Pharmacol. 2014;6:1–17. doi: 10.2147/CPAA.S30338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem S., Mizokami M., Pianko S. NS5A resistance-associated substitutions in patients with genotype 1 hepatitis C virus: prevalence and effect on treatment outcome. J Hepatol. 2017;66:910–918. doi: 10.1016/j.jhep.2017.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Total number of filtered reads obtained for each sample. (B) UPGMA phylogenetic tree of the NS5B segment of 339 nucleotides from the reference sequences used to classify HCV isolates into the 67 subtypes accepted in 2014 (Smith et al., 2014). Bootstrap confidence levels are included on the branches (blue numbers). Genotype 1 references are in dark green, genotype 2 in dark orange, genotype 3 in blue, genotype 4 in pink, genotype 5 in light green, and genotype 6 in yellow.