Figure 5. Association and grouping of sponge symbionts to their HMA/LMA hosts.
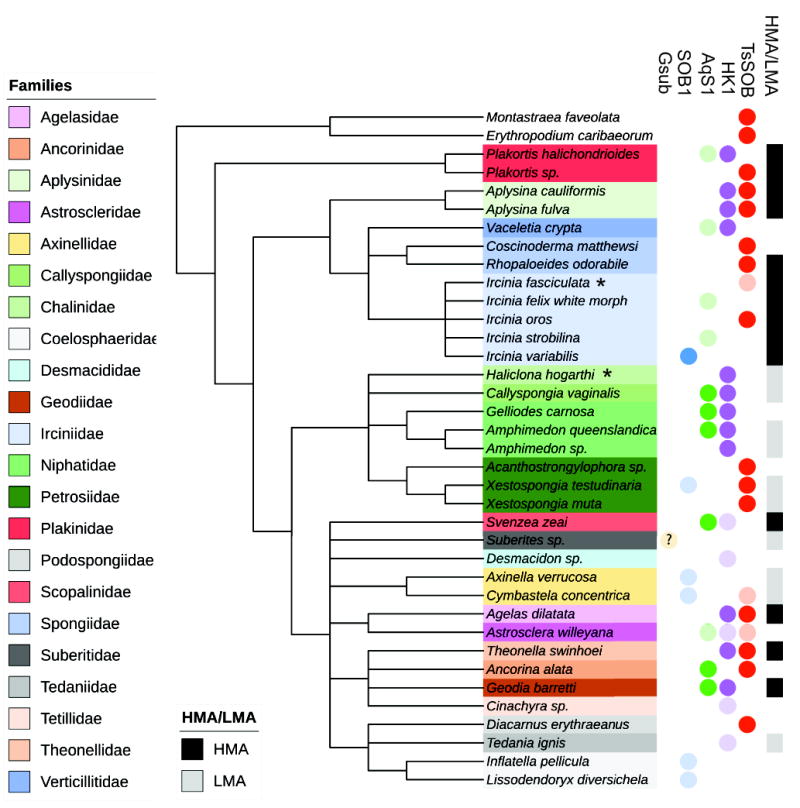
16S rRNA gene of TsSOB is present in multiple orders, and most often in HMA sponges. The 16S rRNA gene of other sulfide oxidizing sponge symbionts (SOB1, AqS1 and HK1, but not Gsub) were also in several sponges. The phylogenetic relations between sponges was adopted from Morrow and Cardenas (2015) and World Porifera Database (Van Soest et al., 2017). The 16S rRNA sequence of Gsub did not match any sponge associated bacterium in the database. The Gsub genome was originally reported from Suberites sp. and therefore it is marked in the tree with a question mark. Sponges were assigned as HMA or LMA according to the works of Gloeckner et al. (2014) and Moitinho-Silva et al. (2015). Lightly colored circles indicate matches that had less than 97% identity and therefor may represent another bacterial species. Two coral species, in which TsSOB was detected, are used as an outgroup. Asterisks denote species names which were updated since the sequences were submitted to NCBI database: Haliclona hogarti is now Haliclona tubifera, Ircinica fasciculata is now Ircinica variabilis, and Ancorinca alata is now Ecionemia alata. The full results of the NCBI nucleotide nr BLAST search are given in Table S3, and the phylogenetic relationship between the 16S rRNA sequences is presented in Figure S9.
