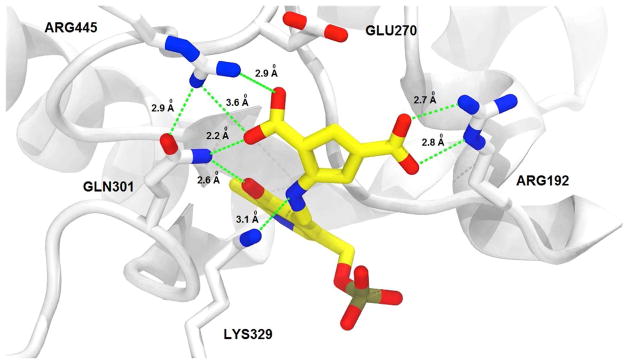
Figure 5.
Structure of GABA-AT inactivated by 5. As with CPP-115, the difluoromethylene group of 5 has been hydrolyzed to a carboxylate moiety. Inactivation of the enzyme appears to be caused by tight binding between the enzyme and a stable PLP-5 noncovalent complex, rather than covalent modification. Color legend: PLP-5 carbons-yellow; protein carbons-white; oxygens-red; nitrogens-blue; phosphorus-taupe. The crystal structure has been deposited in the Protein Data Bank, PDB code 6B6G.