FIG 5.
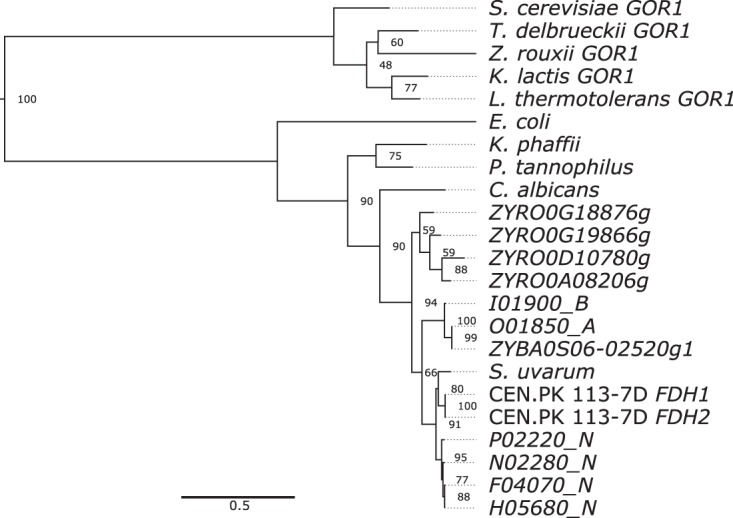
Phylogenetic tree of formate dehydrogenase amino acid sequences in yeast species (including Torulaspora delbrueckii, Zygosaccharomyces rouxii, Kluyveromyces lactis, Lachancea thermotolerans, Komagataella phaffii, Pachysolen tannophilus, Candida albicans, and Saccharomyces uvarum). The six Z. parabailii Fdh-like genes are shown (names ending in _A, _B, and _N). Prefixes ZYRO and ZYBA indicate genes from Z. rouxii and Z. bailii, respectively. The tree was rooted using the paralogous yeast gene GOR1 (glyoxylate reductase), and Escherichia coli FDH is included for reference. The tree was constructed using PhyML. Bootstrap values from 100 replicates are shown.
