Abstract
Previously reported co-occurrence of colorectal cancer (CRC) and tooth agenesis (TA) and the overlap in disease-associated gene variants suggest involvement of similar molecular pathways. Here, we took an unbiased approach and tested genome-wide significant CRC-associated variants for association with isolated TA. Thirty single nucleotide variants (SNVs) in CRC-predisposing genes/loci were genotyped in a discovery dataset composed of 440 individuals with and without isolated TA. Genome-wide significant associations were found between TA and ATF1 rs11169552 (P = 4.36 × 10−10) and DUSP10 rs6687758 (P = 1.25 × 10−9), and positive association found with CASC8 rs10505477 (P = 8.2 × 10−5). Additional CRC marker haplotypes were also significantly associated with TA. Genotyping an independent dataset consisting of 52 cases with TA and 427 controls confirmed the association with CASC8. Atf1 and Dusp10 expression was detected in the mouse developing teeth from early bud stages to the formation of the complete tooth, suggesting a potential role for these genes and their encoded proteins in tooth development. While their individual contributions in tooth development remain to be elucidated, these genes may be considered candidates to be tested in additional populations.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world and a leading cause of cancer-related deaths1. Its etiology is multifactorial and ~33% of all cases are attributed to genetic factors2. Tooth agenesis (TA), the congenital absence of one or more permanent teeth, results from disturbances occurring during tooth development, and represents one of the most common craniofacial anomalies in humans3. Over a decade ago, Lammi et al.4 reported that mutations in the tumor suppressor gene AXIN2 were found co-segregating with colorectal cancer and tooth agenesis in a large multiplex family. Moreover, these authors showed the expression of AXIN2 in developing mouse teeth and suggested that genes involved in colorectal cancer could also be involved in tooth development4.
Tooth development requires a sophisticated series of signaling interactions between the oral epithelium and mesenchyme under strict genetic control by a number of signaling molecules and their downstream signaling pathways5. Any alteration of the epithelial-mesenchymal interactions can have deleterious effects on tooth development, and may affect growth, differentiation and pattern formation, or even have systemic effects6. TA can occur as part of a syndrome, although it is more frequently found as an isolated trait that may appear sporadically or segregating in families, for which the prevalence ranges between ~2–10% excluding third molars. Based on the number of missing teeth, TA is referred to as hypodontia (up to 5 teeth missing), oligodontia (≥6 teeth missing), or anodontia (all teeth missing), the latter being mostly associated with syndromic TA3.
Studies in mice have revealed more than 200 genes involved in tooth development and shown the functional importance of Msx1, Pax9, Pitx2, and Lef1 genes in proper tooth formation and morphogenesis, since the absence of these genes results in arrest of tooth development at the bud stage7–9. In humans, the best characterized mutations for both syndromic and nonsyndromic TA involve MSX1 and PAX97,10, with concordance between mouse and human phenotypes. Additional mutations in genes belonging to the canonical Wnt pathway (i.e., AXIN2, LRP6, WNT10A, WNT10B) have also been increasingly implicated in the susceptibility to nonsyndromic TA11–14, although mouse models have not yet been able to replicate the human phenotypes. Interestingly, Wnt pathway genes have also been shown to play critical roles in tumorigenesis, particularly in colorectal cancer4,15–17.
In addition to epithelial-mesenchymal interactions, cell growth and cell differentiation, the signaling pathways involved in tooth development (i.e., WNT, BMP, SHH, FGF, TGF-β, and NF-kB), also overlap with those implicated in colorectal cancer6,11. Recently, genome-wide association studies (GWAS) have uncovered over 50 loci and single nucleotide polymorphisms (SNPs) that are significantly correlated with the susceptibility to CRC, most of which map to genes with established roles in tumorigenesis, or involved in developmental processes such as transcriptional regulation, genome maintenance, and cell growth and differentiation18–20. Of note, SNPs in CDH1, BMP2, BMP4, and GREM1 genes, known to have important roles in craniofacial and/or tooth development6, have also been reported in association with CRC6,18.
Given the previously reported co-occurrence of CRC and TA and the overlap in disease-associated pathways, we assessed the association of genome-wide associated CRC variants with isolated TA. Here, we describe the association of CRC gene polymorphisms with isolated TA and reveal potentially novel candidate genes for TA in humans and mice.
Results
GWAS-based association study
A total of 440 unrelated Caucasian individuals, 93 with nonsyndromic TA (29 males, 64 females) and 347 unrelated control individuals without TA or family history of TA (101 males, 246 females) were included in this study. All individuals were examined by a dentist, and the presence of tooth agenesis was determined through clinical and radiographic examinations, and considered if one or more permanent teeth were missing from the oral cavity, excluding third molars. Individuals showing signs of syndromic forms of tooth agenesis (i.e., ectodermal dysplasia features) were excluded. Of the 93 cases with TA, 61 presented with hypodontia while 32 presented with oligodontia. Positive family history of cancer was reported by 28 cases and 39 control individuals and included various cancer types including colorectal cancer (Supplementary Table 1).
We selected 30 CRC-predisposing SNVs with genome-wide significance (5 × 10−8) from previously published GWAS19,21,22, for genotyping in our case-control dataset (Table 1). For regions where multiple SNPs have been reported, we included the most significantly-associated SNP according to the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO; https://share.fhcrc.org/sites/gecco/), and prioritized additional SNPs for genotyping based on: (1) location in regulatory or enhancer regions of a given gene, (2) having an effect on motifs in the distal regulatory regions of respective genes, and/or (3) interactions with known genes of biological relevance. Genotypes were generated using Taqman chemistry23 and automatic call rate was >98%.
Table 1.
Details of SNPs investigated in this study.
| Locus | SNP | Base Position | Gene | SNP Location | Allelesa | MAF CEUb |
|---|---|---|---|---|---|---|
| 1q41 | rs6691170 | 221872104 | DUSP10 | downstream | G/T | 0.32 |
| rs6687758 | 221991606 | downstream | A/G | 0.25 | ||
| 2q32.3 | rs11903757 | 191722478 | — | intergenic | T/C | 0.12 |
| 3q26.2 | rs10936599 | 169774313 | MYNN | synonymous coding | C/T | 0.26 |
| 5q31.1 | rs647161 | 135163402 | C5orf66 | intron | C/A | 0.36 |
| 6p21 | rs1321311 | 36655123 | — | intergenic | T/G | 0.29 |
| 8q24 | rs10505477 | 127395198 | CASC8 | intron | T/C | 0.45 |
| rs6983267 | 127401060 | intron | G/T | 0.42 | ||
| rs7014346 | 127412547 | intron | G/A | 0.37 | ||
| 9p24 | rs719725 | 6365683 | — | intergenic | A/C | 0.39 |
| 10p14 | rs10795668 | 8659256 | — | intergenic | G/A | 0.27 |
| rs1665650 | 116727589 | HSPA12A | intron | A/G | 0.28 | |
| 11q13.4 | rs3824999 | 74634505 | POLD3 | intron | A/C | 0.46 |
| 11q23 | rs3802842 | 111300984 | COLCA2 | intron | C/A | 0.29 |
| 12p13.32 | rs10774214 | 4259186 | CCND2 | intron | C/T | 0.39 |
| rs3217810 | 4279105 | intron | C/T | 0.07 | ||
| rs3217901 | 4296223 | intron | A/G | 0.39 | ||
| 12q13.13 | rs7136702 | 50486433 | DIP2B | upstream | T/C | 0.48 |
| rs11169552 | 50761880 | ATF1 | near gene 5' | C/T | 0.27 | |
| 12q24.21 | rs59336 | 114678547 | TBX3 | intron | A/T | 0.47 |
| 14q22.2 | rs4444235 | 53944201 | BMP4 | downstream | T/C | 0.50 |
| rs1957636 | 54093300 | — | intergenic | A/G | 0.50 | |
| 15q13 | rs4779584 | 32702555 | SCG5 | downstream | T/C | 0.24 |
| rs11632715 | 32712046 | GREM1 | upstream | A/G | 0.43 | |
| 16q22.1 | rs9929218 | 68787043 | CDH1 | intron | G/A | 0.26 |
| 18q21 | rs4939827 | 48927093 | SMAD7 | intron | T/C | 0.47 |
| 20p12.3 | rs961253 | 6423634 | — | intergenic | A/C | 0.29 |
| rs4813802c | 6718948 | intergenic | T/G | 0.22 | ||
| 20q13.33 | rs4925386c | 62345988 | LAMA5 | intron | C/T | 0.20 |
| Xp22.2 | rs5934683c | 9783434 | SHROOM2 | upstream | T/C | 0.50 |
aReference allele listed first.
bMAF, minor allele frequency in CEU population (from NCBI dbSNP database).
cOut of Hardy-Weinberg equilibrium and excluded from further analyses.
Genome-wide significant associations were found between TA and individual SNPs on 1q41 and 12q13 (Table 2). At the 1q41 locus, rs6687758 is located downstream of the dual specificity phosphatase 10 (DUSP10) gene, and allelic (P = 2 × 10−9) and genotypic (P = 1.25 × 10−9) associations were found between this SNP and TA, particularly under a recessive model (P = 1.27 × 10−9). At the 12q13.1 locus, rs11169552 is located 2Kb upstream of the activating transcription factor 1 (ATF1) gene promoter, for which genome-wide significant genotypic (P = 4.36 × 10−10) and positive allelic associations (P = 1.87 × 10−7) were also found (Table 2). Positive association was also found between intronic variants in the cancer susceptibility 8 (CASC8) gene on chromosome 8q24 and TA for SNP rs10505477 genotypes and alleles (P = 8.16 × 10−5 and P = 1.7 × 10−5, respectively) and rs7014346 (P = 0.0005 for genotype under a recessive model) (Table 2). When stratifying analyses by cases with hypodontia or oligodontia phenotypes, these associations remained significant for both phenotypes, despite the smaller number of oligodontia cases (P ≤ 0.002, data not shown). To confirm our findings, we genotyped the associated SNPs on an independent dataset composed of 52 cases with tooth agenesis and 427 gender- and ethnicity-matched unrelated controls, and found significant association between CASC8 rs10505477 (P = 0.006 for genotype and P = 0.008 for allele) and TA, particularly under a dominant model (P = 0.001).
Table 2.
Summary of association results under different genetic test models.
| Gene | SNP Id. | MAF CEUa | MAF Case | MAF Control | Test | Alleles | Frequency Cases | Frequency Controls | P-valueb |
|---|---|---|---|---|---|---|---|---|---|
| ATF1 | rs11169552 | Genotype | CC/CT/TT | 38/31/24 | 195/129/14 | 4.36 × 10 −10 | |||
| 0.27(T) | 0.42(T) | 0.24(T) | Allele | C/T | 107/79 | 519/157 | 1.85 × 10−7 | ||
| Dominant | CC × CT + TT | 38/55 | 195/143 | 0.004 | |||||
| Recessive | CC + CT × TT | 69/24 | 324/14 | 6.79 × 10 −11 | |||||
| DUSP10 | rs6687758 | Genotype | AA/AG/GG | 37/34/22 | 217/112/14 | 1.25 × 10 −9 | |||
| 0.25(G) | 0.42(G) | 0.20(G) | Allele | A/G | 108/78 | 546/140 | 2.09 × 10 −9 | ||
| Dominant | AA × AG + GG | 37/56 | 217/126 | 5.09 × 10−5 | |||||
| Recessive | AA + AG × GG | 71/22 | 329/14 | 1.27 × 10−9 | |||||
| CASC8 | rs10505477 | Genotype | TT/TC/CC | 16/45/32 | 119/167/56 | 8.16 × 10−5 | |||
| 0.45(C) | 0.41(T) | 0.41(C) | Allele | T/C | 77/109 | 405/279 | 1.69 × 10−5 | ||
| Dominant | TT × TC + CC | 16/77 | 119/223 | 0.001 | |||||
| Recessive | TT + TC × CC | 61/32 | 286/56 | 0.0001 | |||||
| rs7014346 | Genotype | GG/GA/AA | 31/38/24 | 145/154/39 | 0.002 | ||||
| 0.37(A) | 0.46(A) | 0.34(A) | Allele | G/A | 100/86 | 444/232 | 0.003 | ||
| Dominant | GG × GA + AA | 31/62 | 145/193 | 0.09 | |||||
| Recessive | GG + GA × AA | 69/24 | 299/39 | 0.0005 |
MAF, minor allele frequency.
aReference allele.
bFisher Exact test, denotes genome-wide significant association if P ≤ 5 × 10−8 (bolded) and positive association under Bonferroni correction if P ≤ 0.002 (italicized).
Significant haplotype associations were also observed and included the individually associated SNPs. The strongest association was observed for the combination of CASC8 rs10505477, rs7013436, and rs6983267 alleles (CAG haplotype, P = 5 × 10−13; CAT haplotype, P = 4 × 10−9), followed by DUSP10 rs6687758 and rs6691170 (GG haplotype, P = 7.32 × 10−10), and ATF1 rs7136702 and rs11169552 (TC haplotype, P = 4.8 × 10−8) (Supplementary Table 2). Interestingly, when considering TA in the presence of family history of cancer, positive association was found for carriers of a TT genotype (thus two copies of the minor allele T) in ATF1 rs11169552 (P = 0.00004), and for carriers of a GG genotype in DUSP10 rs6687758 (P = 0.003) (Supplementary Table 3).
Functional annotation of associated variants
As a first step into assessing the relevance of the associated genes in tooth agenesis phenotypes, we annotated the associated variants using the dbSNP24 and the UCSC Genome Browser25 databases and to predict gene and variant function. Our analyses showed that the associated variants, although common, are predicted to have effects on chromatin structure and RNA polymerase activity and on transcription of their respective genes. ATF1 rs11169552 is located in the gene promoter in a region showing high DNase I hypersensitivity clusters and potential location of regulatory elements (enhancers, silencers, insulators, and locus control region) (Supplementary Figure 1). Further, ATF1 rs11169552 is predicted to harbor a binding site for POLR2A, essential for RNA polymerase activity and DNA transcription. DUSP10 rs6687758 is located in a DNAse I hypersensitivity cluster with binding motif for TBP (TATA-box binding protein) (Supplementary Figure 2). CASC8 rs10505477 appears to harbor a putative binding motif to NK2–5, in a region of enriched H3K27Ac histone marks (Supplementary Figure 3). These findings suggest that these variants are likely to have functional consequences on gene expression.
We attempted to presume the effects of the associated SNPs on expression of their respective genes by searching for evidence of eQTLs at the respective loci, however no data was available in relevant oral/dental tissues to yield conclusive evidence (data not shown).
Expression analyses
Since no information was available from public databases regarding the expression of ATF1, DUSP10 and CASC8 during tooth development, as a second step into determining the relevance of these genes in tooth development, we sought to assess whether their transcripts and/or encoded proteins were expressed in the oral and dental tissues of developing mouse embryos.
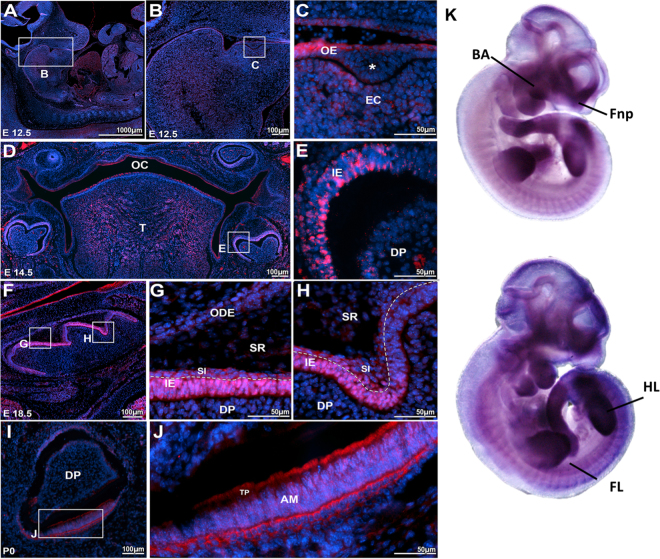
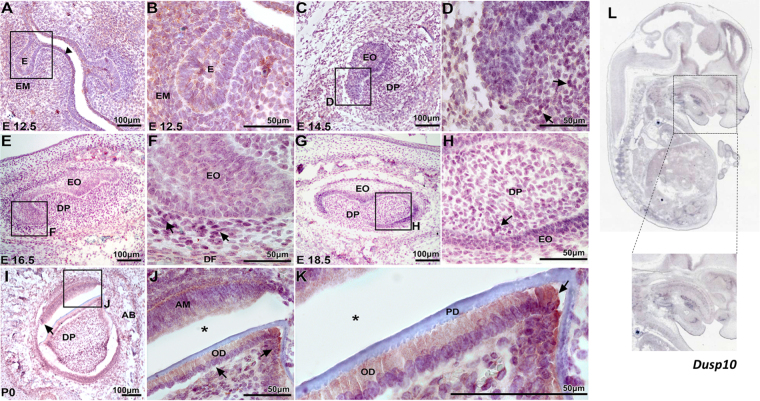
We performed immunofluorescence and immunohistochemistry procedures to detect the localization of Atf1 and Dusp10, respectively, during murine tooth development stages. At E12.5, Atf1 expression was detected in the oral epithelial cells and in the subjacent condensed ectomesenchymal cells (Fig. 1A,C). Later, at E14.5, Atf1 expression was evident in the oral and tongue epithelia, and underlying mesenchyme, in the inner dental epithelium (IE) and in few scattered cells of the dental papilla (DP) (Fig. 1D–E). At E16.5 (Supplementary Figure 4), and E18.5, Atf1 was markedly expressed in the inner epithelium and the stratum intermedium. Reduced expression was also noted in the cells from the stratum intermedium, stellate reticulum and outer dental epithelium (Fig. 1F,H). At P0, Atf1 expression shifted to the cytoplasm of the polarized layer of ameloblasts, in a perinuclear pattern in the ameloblast bodies, as well as in the Tomes’ processes (Fig. 1I,J). Atf1 mRNA expression had been shown to be expressed in mouse embryos as early as E10.5, with strong expression noted in the mesenchyme of frontonasal prominences, branchial arches and limbs26 (Fig. 1K). Dusp10 expression was detected at a site of proliferation of the dental placode at E12.5, surrounded by a condensation of the underlying ectomesenchymal cells compatible with a bud stage of tooth development. Expression was evident in the epithelial cells from the oral epithelium, as well as in the condensed ectomesenchymal cells surrounding the epithelial proliferation (Fig. 2A,B). At E14.5, nuclear and cytoplasmic staining was observed on the cells from the enamel organ and dental papillae (Fig. 2C,D). At bell stage E16.5, positive staining was observed in the enamel organ, dental papillae and dental follicle cells (Fig. 2E,F). At 18.5, Dusp10 expression was noted in the enamel organ, in the ectomesenchymal cells of the dental papillae and the dental follicle cells (Fig. 2G,H). Later, at P0, Dusp10 expression was localized to the preameloblasts, odontoblasts and pre-dentin (Fig. 2I,K, Supplementary Figure 5). Marked expression was noted in the nuclei and cytoplasms of preameloblasts, as well as in the subjacent odontoblast layer (Fig. 2K). Dusp10 mRNA expression was observed in the mouse craniofacial region and oral cavity at E14.5, particularly in the mouth and tongue27. (Fig. 2L). These data suggested that Atf1 and Dusp10 are present throughout tooth development stages in mouse embryos and thus likely to have a role in tooth development. Therefore, variations that impact the function of ATF1 and DUSP10 in humans are likely to play a contributory role in tooth agenesis.
Figure 1.
Atf1 is expressed in mouse developing teeth. (A,B) Sagittal section of mouse embryo at E12.5. (C) Proliferation (*) of the oral epithelium (OE) into the ectomesenchyma, (EM) consistent with the formation of a molar tooth bud at early bud stage. ATF1 expression was detected the oral epithelium, as well as in the condensed ectomesenchymal cells. (D) Panoramic photomicrography showing the mouse oral cavity (OC) with tooth germs at early bell stage (E14.5). (E) ATF1 expression was noted in the inner enamel/dental epithelium (IE) and dental papilla (DP). (F–H) At E16.5 (Appendix Figure 1) and E18.5, marked ATF1 expression was observed in the inner enamel epithelium (IE) and in the stratum intermedium (SI), with sparse expression noted in the stellate reticulum (SR) and outer dental (ODE) epithelium. (I–J) In incisor teeth at P0, ATF1 expression was particularly evident in the polarized layer of ameloblasts, and in the Tomes’ processes (TP). (K) Atf1 mRNA detected by whole mount in situ hybridization with digoxigenin-labeled antisense RNA followed by alkaline phosphatase-coupled antibody against digoxigenin in C57BL mice at embryonic day 10.5. Strong expression is noted in the mesenchyme of frontonasal prominences, branchial arches and limbs (Obtained from MGI Gene Expression Database. Original source: Gray et al. Mouse Brain Organization Revealed Through Direct Genome-Scale TF Expression Analysis. Science. 2004 24;306(5705): 2255–2257). Secondary antibody goat anti-rabbit-Alexa 555 for detection of ATF1 and DAPI for nuclear staining. OE = Oral ephitelium; EM = ectomesenchymal cells; OC = oral cavity; T = tongue; IE = inner epithelium; DP = dental papilla; SI = stratum intermedium; SR = stellate reticulum; ODE outer dental ephithelium; AM = ameloblasts; TP = Tomes’ processes; BA = branchial arches; Fnp = frontonasal processes; HL = hind limbs; FL = forward limbs.
Figure 2.
Dusp10 expression in mouse developing teeth. (A–B) At E12.5, there is a proliferation of the dental lamina surrounded by a condensation of the underlying ectomesenchymal cells (EM) compatible with a bud stage of tooth development. Dusp10 expression was noted in the epithelial cells from oral epithelium (arrowhead), as well as in the condensed ectomesenchymal cells surrounding the epithelial proliferation (E). (C,D) At E14.5, expression was noted detected in the enamel organ (EO) and dental papillae (DP) (arrows). (E,F) At bell stage E16.5, Dusp10 positive staining is observed in the enamel organ, dental papillae and dental follicle (DF) cells (arrows). (G,H) At E18.5, the tooth germ demonstrates morphodifferentiation compatible with a bell stage. The enamel organ (EO) cells, ectomesenchymal cells of the dental papillae (DP) and the dental follicle cells show homogenous nuclear immune staining. (I–K) At P0, the incisor tooth germ is surrounded by alveolar bone (AB) and shows morphology compatible with a late bell stage or crown stage, such as ameloblasts (AM), odontoblasts (OD) and pre-dentin (PD) deposition. Dusp10 expression was observed in the preameloblasts and subjacent odontoblast layer (arrows). (L) Dusp10 mRNA detected by whole mount in situ hybridization in C57BL/6 mouse at E14.5. Positive expression is noted in the craniofacial region and oral cavity, particularly in the mouth and tongue. (Obtained from MGI Gene Expression Database. Original source: Hoffman et al. Genome Biol 2008;9(6):R99). An artifact (*) separated the preameloblasts from the pre-dentin and odontoblastic layer. DAB chromogen and counterstaining with Mayer’s Hematoxylin. OE = Oral ephitelium; EM = ectomesenchymal cells; EO = enamel organ; DP = dental papilla; AM = ameloblasts; AB = alveolar bone.
CASC8 has only recently been annotated in the human genome, and not yet in the mouse genome, and thus limited our analysis to evaluate its expression in relevant tissues. Using information available in public databases, we found that CASC8 mRNA is expressed in many tissues including brain, skin, minor salivary gland, colon and esophagus. We then performed RT-PCR analysis of CASC8 expression in stem cells of the apical papilla (SCAP), a dental-derived stem cell line28, however, no expression was detected.
Discussion
Over a decade ago, Lammi et al.4 identified AXIN2 as a novel gene involved in tooth development after finding mutations in this gene to be co-segregating with CRC and TA in a large multiplex family. Moreover, these authors showed the expression of Axin2 in developing mouse teeth and showed for the first time that a gene involved in CRC could also be involved in tooth development. Here, we describe the findings of CRC-predisposing genes with potential roles in tooth development and as new candidate genes for isolated TA.
From the thirty CRC-predisposing variants assessed, our association analyses revealed genome-wide significant associations between TA and loci on 1q41 and 12q13, containing DUSP10 (dual specificity phosphatase 10) and ATF1 (activating transcription factor 1) genes, respectively. Meanwhile, positive associations (although not strong to reach the genome-wide significance threshold level) were also found between CASC8 (cancer susceptibility candidate 8) and TA. These genes had not yet been described to have a role in tooth development and prompted investigations regarding their biological relevance to the TA phenotype. Intriguingly, no associations were found between TA and CDH1, BMP2, BMP4, or GREM1 genes, known to have important roles in craniofacial and/or tooth development6, and which have also been reported in association with CRC6,18.
ATF1 belongs to the ATF/CREB family of transcription factors, which binds to the consensus ATF/CRE site ‘TGACGTCA’ and regulates the transcription of target genes to participate in various cellular processes29. Previous reports showed that the function of ATF1 appears to depend on the cellular and genetic context to play an important role in tumor progression in a tumor-specific manner30. ATF1 was found to be over-expressed in several cancer types including lymphoma, melanoma and nasopharyngeal carcinoma, and functioned as a tumor promoter both in vitro and in vivo31–34. In contrast, disruption of ATF1 activity suppressed its tumorigenicity and metastatic potential33. However, the exact role of ATF1 during craniofacial development is still largely unknown. Loss of Atf1 in mice did not cause any obvious phenotypic abnormalities although led to increased programmed cell death35.
DUSP10 is a dual specificity phosphatase (DUSP) that negatively regulates the activation of the mitogen-activated protein kinase (MAPK) gene family, which includes the extracellular-regulated kinases (ERKs) and the stress-activated protein kinases p38 and c-Jun NH2-terminal kinase (JNK). Activation of MAPK by various stimuli including growth factors, cytokines, or stress conditions, regulates major cellular responses such as proliferation, differentiation, survival, migration or production of soluble factors36. Dusp10 has been regarded as an important regulator of tumorigenesis in animal models37, and loss of Dusp10 in mice caused enhanced immune and inflammatory responses although effects on skeletal phenotype have not been described38. Nonetheless, other DUSPs have been found to play critical roles in development. Loss of function studies revealed that Dusp4 is essential for early development and endoderm specification in the zebrafish39, whereas Dusp5 was described to control angioblast populations in the lateral plate mesoderm40.
CASC8 is a long noncoding RNA (lncRNA), located in the gene desert region on 8q24.21. LncRNA is a new class of transcripts that are involved in multiple cellular functions including the regulation of expression of multiple genes. Studies have shown that SNPs in lncRNAs may affect the biological processes of messenger RNA conformation, and results in the modification of its interacting partners. Interestingly, various cancer types, including CRC, prostate, breast and gastric cancer, have been reported in association with CASC822,41, possibly through its function as a lncRNA.
Variants in ATF1, DUSP10 and CASC8 have been associated with increased risk of CRC in different GWAS with multiple populations19,21,22, although functional studies addressing the potential effects of these variants on gene function and/or disease mechanisms are scarce. While the biological function of the associated ATF1, DUSP10, and CASC8 variants is yet to be determined, our bioinformatics analyses revealed that all three variants are predicted to be within DNAse I hypersensitivity clusters, or in enhancer histone marks, with likely functional effects on gene expression. Moreover, ATF1 rs11169552 is predicted to harbor a binding site for POLR2A, essential for RNA polymerase activity and DNA transcription. Another regulatory variant in ATF1 (rs11169571) was shown to increase breast/ovarian cancer risk through modifying miRNA binding42,43. Interestingly, this variant is in linkage disequilibrium with ATF1 rs11169552, associated in the present study, and could be transmitting the same genetic information as well as functional potential. DUSP10 rs6687758 is predicted to have a putative binding motif for TBP (TATA-box binding protein), which controls the transcription machinery. CASC8 rs10505477 has a putative binding motif to NK2-5, a homeobox-containing transcription factor with roles in embryonic development. These findings suggest that these genes are active in early cell processes being important in both embryogenesis and tumorigenesis.
The relevance of ATF1, DUSP10 and CASC8 in tooth development is unknown. Therefore, we assessed the expression of these genes or their encoded proteins in relevant tissues or cell lines. During mouse embryonic development, Atf1 mRNA was detected at embryonic day 10.5, with strong expression noted in the mesenchyme of the frontonasal prominences, branchial arches and limbs26. Our findings showed that Atf1 is expressed throughout mouse tooth development stages, in the oral epithelial cells and subjacent condensed ectomesenchymal cells during the early bud stage of tooth development, then shifting to the inner dental epithelium and dental papilla, and finally in the ameloblasts at the final stages of tooth development. Dusp10 mRNA expression was also observed in the mouse craniofacial region and oral cavity at E14.5, particularly in the mouth and tongue27. In our study, the expression of Dusp10, albeit weak, was also noted in the proliferating dental tissues at the early stages of tooth development. At later stages, Dusp10 expression was evident in the enamel organ, pre-ameloblasts and odontoblastic layer. Expression of CASC8 mRNA has been observed in the brain, skin, minor salivary gland, colon and esophagus. However, no expression of CASC8 was detected in our study with human dental stem cells of the apical papilla (SCAP). Since CASC8 has only recently been annotated in the human genome (and not yet annotated in mice), we were limited in the choice of relevant tissues to assess its expression thus hampering our interpretation of the role of this gene in tooth development. Of note, the observed association between TA and the CASC8 locus on chromosome 8q24.21 raises intriguing questions as this locus has been associated with increased risk of several malignancies including CRC22,41 and also cleft lip/palate44,45, for which an expanded phenotype including tooth agenesis has been proposed46. Furthermore, the associated CASC8 rs10505477, is located at approximately 250–300 Kb away from the transcription start site of cMYC, a gene well-known for its reprogramming capacity and as a proto-oncogene. Enhancer elements of cMYC are known to be up to 500 Mb away from the gene start site, therefore it is possible that the CASC8 SNP may be in a LD block associated with cMYC enhancer elements and drive the association findings47. The presence of Myc has been detected in dental pulp cells, dental follicle cells, ameloblasts and odontoblasts, and could be related to cell reprogramming or differentiation during tooth development48.
Among the limitations of this study is the limited availability of individual cancer history or family history of cancer. Since most of our patients with TA are relatively young, our individual patient tumor/cancer history was low (3 patients). In addition, the majority of the TA patients’ parents and/or immediate family members are also younger than the ages in which preventive cancer screening begins. Regarding family history of cancer, in some instances, patients were unsure of cancer status in the family and thus their responses were not accounted for. We considered positive family history of cancer when the individual could name the relative that was affected, and despite various cancer types (including colorectal cancer) were reported for relatives of TA patients, the number of cases was considered small to warrant conclusive genotype-phenotype comparisons between cases and controls. It is also possible that the genetic variants associated with TA in the present study may be in linkage disequilibrium with a more distant causal variant, and acting as surrogate markers for the condition. Additional fine-mapping around the associated loci may provide additional insights into the association signal. Lastly, we cannot rule out that additional variants in other genes may also be contributing to the TA phenotype observed. Similar to other genetic conditions, the hypothesis of multilocus genomic variation49 was proposed for some families in which a single genetic variant could not explain the TA phenotypes13. In this context, the association observed with the CRC-predisposing genes may represent a direct or indirect role in the background of additional causal genes.
In summary, our findings provide evidence that novel genes and gene pathways may be yet unknown for a role in tooth development, and unbiased genetic studies have the potential to identify the full spectrum of candidate genes and variants associated with isolated TA. While the exact contributions of ATF1, DUSP10 and CASC8 in TA remain to be further elucidated, our findings further support that genes involved in colorectal cancer may also be involved in tooth development and provide additional insights into deciphering the complex etiology of the condition.
Methods
Study Population
This study was approved by the University of Texas Health Science Center at Houston Committee for the Protection of Human Subjects and the University of Pittsburgh Institutional Review Board, and all methods were performed in accordance with the relevant guidelines and regulations. Unrelated individuals with and without tooth agenesis were invited to participate in the study when presenting for treatment at the University of Texas Health Science Center at Houston School of Dentistry clinics. Written informed consent was obtained from all participating individuals and/or from their parents or guardians. Clinical and demographic information, and saliva samples were collected from all individuals. When known, individual and familial history of cancer was also recorded. All individuals were examined by a dentist, and the presence of tooth agenesis was determined through clinical and radiographic examinations, and considered if one or more permanent teeth were missing from the oral cavity, excluding third molars. Differential diagnosis of tooth agenesis from other potential causes of tooth loss (e.g. caries, periodontal disease, trauma) was ensured through comprehensive evaluation of patients’ electronic dental records and conversation with the patient and/or legal guardian about previous tooth extractions and history of trauma. Individuals showing signs of syndromic forms of tooth agenesis (i.e., ectodermal dysplasia features) were excluded.
To avoid potential effects of population stratification in genetic studies, only individuals with self-reported Caucasian ethnicity (up to 2 generations) were included in the study. A total of 440 unrelated individuals, 93 with nonsyndromic TA (29 males, 64 females) and 347 ethnicity-matched, control individuals without TA or family history of TA (101 males, 246 females) were included in this study. No selection regarding gender was used; the female to male ratio observed in this study is similar to the overall gender prevalence reported in the literature.
Of the 93 cases with TA, 61 presented with hypodontia while 32 presented with oligodontia. Positive family history of cancer was reported by 28 cases and 39 control individuals. Details of known cancer types reported are available in Supplementary Table 1.
Selection of CRC-Associated Variants and Genotyping
We selected 30 CRC-risk-associated SNPs that reached genome-wide significance (5 × 10–8) in previously published GWAS19,21,22 to genotype in this study (Table 2). We prioritized the SNPs to investigate in our study, based on the population that they had been investigated to more closely match our study population, and genome-wide significant P-values. For regions where multiple SNPs have been reported, we included the most significantly-associated SNP according to the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO; https://share.fhcrc.org/sites/gecco/), and prioritized additional SNPs for genotyping based on: (1) location in regulatory or enhancer regions of a given gene, (2) having an effect on motifs in the distal regulatory regions of respective genes, and/or (3) interactions with known genes of biological relevance.
Genotyping was performed using Taqman chemistry23 in 5-μL final reaction volumes in a ViiA7 Sequence Detection System (Applied Biosystems, Foster City, CA). Results were analyzed using EDS v.1.2.3 software (Applied Biosystems). For quality control of genotyping reactions, we used a non-template reaction as negative control and a DNA sample of known genotype as positive control. Genotyping was performed in duplicates and genotypes that did not have a 95% pass rate were repeated.
Data analyses
Power calculations were performed using Genetic Power Calculator (http://zzz.bwh.harvard.edu/gpc/) and indicate that the study sample size provided approximately 86% power to detect an association with an alpha of 0.05, if the markers selected are in linkage disequilibrium with the causal factor (D′ = 0.80) and their frequencies are around 80%. Data analysis was performed using PLINK version 1.0650. Hardy-Weinberg equilibrium was calculated for cases and controls, and SNPs showing evidence of deviation in controls were excluded from further analyses. Differences in allele and genotype frequencies for each polymorphism between cases and controls were compared using chi-square and Fisher Exact tests. We corrected for multiple testing using the Bonferroni method, and the significance level was set considering the number of tests (n = 30) to give a corrected P-value (α = 0.002). Haplotype analyses were performed using the ‘haplotype-based case-control association test’ as implemented in PLINK. Regression analyses were performed using EpiInfo v.7.1 (https://www.cdc.gov/epiinfo/) to identify potential preferential associations between specific SNP genotypes with tooth agenesis adjusted for family history of cancer and gender.
Bioinformatic analyses of SNP function
Annotation of the newly identified loci was performed using the dbNSFP24 and FuncPred (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html) databases. Information on the function of the associated SNPs regarding splicing regulation, stop codon, Polyphen predictions, transcription factor binding site predictions, miRNA binding site prediction regulatory potential score, conservation score, and nearby genes, were recorded. The presence of expression quantitative trait loci for each of the associated SNPs was assessed using the GTEx portal (gtexportal.org).
Immunofluorescence and immunohistochemistry
The expression of Atf1 and Dusp10 proteins was evaluated using immunofluorescence and immunohistochemistry in developing mouse tooth sections, respectively. Wild type C57BL/6 mouse embryos at embryonic days (E) 12.5, 14.5, 16.5, 18.5 and postnatal day 0 (P0) were harvested, fixed in 4% paraformaldehyde and embedded in paraffin for sectioning at 7μm of thickness. Sections were deparaffinized in xylene solution, rehydrated in gradative alcohol baths and rinsed with deionized H2O at room temperature. For antigen retrieval, sections were immersed in sodium citrate buffer (10 mM, pH 6) at 100 °C for 30 minutes and allowed to cool down for 30 min at room temperature. To avoid nonspecific binding of the antibodies, sections were immersed in blocking solution (1% bovine serum albumin and 10% goat serum diluted inPBS) and incubated with AffiniPure Fab fragment goat anti-mouse (Jackson ImmunoResearch Laboratories, PA, USA), for 1 hour at room temperature. For Dusp10, an additional blocking step consisted of incubating sections in 2.5% normal horse serum (Vector Laboratories, CA, USA) for 30 min at room temperature. Sections were then washed and incubated with either Atf1 (rabbit polyclonal anti-mouse Atf1; ab189311, Abcam, MA, USA), or Dusp10 (rabbit polyclonal anti-mouse Dusp10; ab71309, Abcam, MA, USA) primary antibodies at 4 °C overnight. For Atf1, sections were then washed and incubated with Alexa Fluor® 555 (goat anti-rabbit secondary antibody, A-21428, ThermoFisher Scientific,MA, USA) for 2 hours in a dark chamber at room temperature. Lastly, sections were washed (3 × 10′), counterstained with DAPI (Invitrogen) in a dark chamber for 10 min, and mounted with ProLong Gold AntifadeReagent (Invitrogen). For Dusp10, sections were incubated with biotinylated horse anti-rabbit IgG (BP-1100, Vector Laboratories, CA, USA) for 30 minutes at room temperature, washed and incubated with Streptavidin/Peroxidase complex (PK7800, Vector Laboratories, CA, USA) for 5 minutes and washed. Sections were then incubated in 3,3′-Diaminobenzidine (ImPACT DAB, Vector Laboratories, CA, USA) chromogen for 2 minutes, washed, and counterstained with Mayer’s Hematoxylin. Lastly, sections were washed, dehydrated and mounted. Imaging was performed after 48 hours in a Nikon Eclipse Ni-U upright fluorescence microscope (Nikon, Germany) equipped with a Zyla 5.5 sCMOS camera (Andor).
Reverse transcriptase-PCR (RT-PCR)
Total RNA was extracted from SCAP by using the RNeasy kit (Qiagen Inc, Valencia, California, USA). RNA sample integrity was checked by analyzing 1 μg of total RNA on 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California, USA). After RNA extraction, complementary DNA was synthesized by using 3 μg of RNA through a reverse transcription reaction using QuantiTectRT kit (Qiagen Inc, Valencia, California, USA). Four sets of primers for CASC8 amplification were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) (Supplementary Table 4). β-actin (Actb) was used as endogenous control. PCR reactions were performed in 25 uL final reaction volume using 20 ng of SCAP cDNA, 1uL of each forward and reverse primers, 0.5 uL Taq DNA polymerase, MgCl2, deoxynucleotide mix, 10 × PCR buffer, and water. Reaction conditions were: 40 cycles at 95 °C (10′), 94 °C (1′), 56 °C (1′), and 72 °C (2′). Products were resolved in agarose gel electrophoresis and imaged using Odyssey Scanner (LI-COR Biosciences, Lincoln, NE).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank the study individuals for participation in this study. This work was supported in part by funding from the National Institute for Dental and Craniofacial Research (NIDCR) (R03-DE024596 to A.L., and R90 DE024296-03 to M.R.B.); American Association of Orthodontists (Foundation Biomedical Research Award to S.A. and A.L.); the Rolanette and Berdon Lawrence Bone Disease Program of Texas (to W.D.F.), and the Scientific and Technological Research Institution of Turkey, TUBITAK-ERA NET (CRANIRARE-2, grant number: SBAG-112S398), Istanbul University Research Fund (Project No: 48398). M.A.W. was supported by the UTSD Summer Research Program. Data and samples from Pittsburgh were sourced through the Dental Registry and DNA Repository project, which is supported by the University of Pittsburgh School of Dental Medicine.
Author Contributions
M.A.W., contributed to data acquisition, analyses and interpretation, drafted the manuscript. C.B., M.R.B., contributed to data acquisition, analyses and interpretation, critically revised the manuscript. K.M., F.C., N.D., X.L., R.S., S.A., A.R.V., B.A.A., W.D.F., contributed to data analyses and interpretation, critically revised the manuscript. A.L. contributed to study conception, design, data analyses and interpretation, drafted the manuscript, critically revised the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21368-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Navarro M, Nicolas A, Ferrandez A, Lanas A. Colorectal cancer population screening programs worldwide in 2016: An update. World J Gastroenterol. 2017;23:3632–3642. doi: 10.3748/wjg.v23.i20.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 3.Hennekam, R. C. K. I. D. & Allanson, J. E. Gorlin’s Syndromes of the Head and Neck. 5th edn, (Oxford University Press, 2010).
- 4.Lammi L, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. American journal of human genetics. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997;67:111–123. doi: 10.1016/S0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- 6.Cudney SM, Vieira AR. Molecular factors resulting in tooth agenesis and contemporary approaches for regeneration: a review. European archives of paediatric dentistry: official journal of the European Academy of Paediatric Dentistry. 2012;13:297–304. doi: 10.1007/BF03320830. [DOI] [PubMed] [Google Scholar]
- 7.Thesleff I, Vaahtokari A, Vainio S, Jowett A. Molecular mechanisms of cell and tissue interactions during early tooth development. The Anatomical record. 1996;245:151–161. doi: 10.1002/(SICI)1097-0185(199606)245:2<151::AID-AR4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 8.Lan Y, Jia S, Jiang R. Molecular patterning of the mammalian dentition. Semin Cell Dev Biol. 2014;25-26:61–70. doi: 10.1016/j.semcdb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CR, et al. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 10.Li X, et al. A model for the molecular underpinnings of tooth defects in Axenfeld-Rieger syndrome. Hum Mol Genet. 2014;23:194–208. doi: 10.1093/hmg/ddt411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin W, Bian Z. The Gene Network Underlying Hypodontia. Journal of dental research. 2015;94:878–885. doi: 10.1177/0022034515583999. [DOI] [PubMed] [Google Scholar]
- 12.Massink MP, et al. Loss-of-Function Mutations in the WNT Co-receptor LRP6 Cause Autosomal-Dominant Oligodontia. American journal of human genetics. 2015;97:621–626. doi: 10.1016/j.ajhg.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinckan N, et al. Whole-Exome Sequencing Identifies Novel Variants for Tooth Agenesis. J Dent Res. 2018;97:49–59. doi: 10.1177/0022034517724149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu P, et al. Mutations in WNT10B Are Identified in Individuals with Oligodontia. American journal of human genetics. 2016;99:195–201. doi: 10.1016/j.ajhg.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin W, Bian Z. Hypodontia, a prospective predictive marker for tumor? Oral Dis. 2016;22:265–273. doi: 10.1111/odi.12400. [DOI] [PubMed] [Google Scholar]
- 16.Letra A, Menezes R, Granjeiro JM, Vieira AR. AXIN2 and CDH1 polymorphisms, tooth agenesis, and oral clefts. Birth defects research. Part A, Clinical and molecular teratology. 2009;85:169–173. doi: 10.1002/bdra.20489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan N, et al. Axis inhibition protein 2 (AXIN2) polymorphisms and tooth agenesis. Archives of oral biology. 2009;54:45–49. doi: 10.1016/j.archoralbio.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nature genetics. 2014;46:533–542. doi: 10.1038/ng.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu L, et al. A model to determine colorectal cancer risk using common genetic susceptibility loci. Gastroenterology. 2015;148:1330–1339 e1314. doi: 10.1053/j.gastro.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang BW, et al. Association between GWAS-identified genetic variations and disease prognosis for patients with colorectal cancer. PLoS One. 2015;10:e0119649. doi: 10.1371/journal.pone.0119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houlston RS, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemire M, et al. A genome-wide association study for colorectal cancer identifies a risk locus in 14q23.1. Hum Genet. 2015;134:1249–1262. doi: 10.1007/s00439-015-1598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranade K, et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–1268. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Wu C, Li C, Boerwinkle E. dbNSFPv3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent, W. J. et al. The human genome browser at UCSC. Genome research12, 996–1006, 10.1101/gr.229102. Article published online before print in May2002 (2002). [DOI] [PMC free article] [PubMed]
- 26.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman BG, et al. Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biol. 2008;9:R99. doi: 10.1186/gb-2008-9-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Gronthos S, Shi S. Dental pulp stem cells. Methods Enzymol. 2006;419:99–113. doi: 10.1016/S0076-6879(06)19005-9. [DOI] [PubMed] [Google Scholar]
- 29.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/S0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 30.Huang GL, et al. Activating transcription factor 1 is a prognostic marker of colorectal cancer. Asian Pac J Cancer Prev. 2012;13:1053–1057. doi: 10.7314/APJCP.2012.13.3.1053. [DOI] [PubMed] [Google Scholar]
- 31.Hsueh YP, Lai MZ. Overexpression of activation transcriptional factor 1 in lymphomas and in activated lymphocytes. J Immunol. 1995;154:5675–5683. [PubMed] [Google Scholar]
- 32.Huang GL, et al. The protein level and transcription activity of activating transcription factor 1 is regulated by prolyl isomerase Pin1 in nasopharyngeal carcinoma progression. Cell Death Dis. 2016;7:e2571. doi: 10.1038/cddis.2016.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jean D, Bar-Eli M. Regulation of tumor growth and metastasis of human melanoma by the CREB transcription factor family. Mol Cell Biochem. 2000;212:19–28. doi: 10.1023/A:1007128101751. [DOI] [PubMed] [Google Scholar]
- 34.Su B, et al. Stage-associated dynamic activity profile of transcription factors in nasopharyngeal carcinoma progression based on protein/DNA array analysis. OMICS. 2011;15:49–60. doi: 10.1089/omi.2010.0055. [DOI] [PubMed] [Google Scholar]
- 35.Bleckmann SC, et al. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol Cell Biol. 2002;22:1919–1925. doi: 10.1128/MCB.22.6.1919-1925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bermudez O, Pages G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299:C189–202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 37.Png CW, et al. DUSP10 regulates intestinal epithelial cell growth and colorectal tumorigenesis. Oncogene. 2016;35:206–217. doi: 10.1038/onc.2015.74. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature. 2004;430:793–797. doi: 10.1038/nature02764. [DOI] [PubMed] [Google Scholar]
- 39.Brown JL, et al. Transcriptional profiling of endogenous germ layer precursor cells identifies dusp4 as an essential gene in zebrafish endoderm specification. Proc Natl Acad Sci USA. 2008;105:12337–12342. doi: 10.1073/pnas.0805589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pramanik K, et al. Dusp-5 and Snrk-1 coordinately function during vascular development and disease. Blood. 2009;113:1184–1191. doi: 10.1182/blood-2008-06-162180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao K, Hua L, Wei L, Meng J, Hu J. Correlation Between CASC8, SMAD7 Polymorphisms and the Susceptibility to Colorectal Cancer: An Updated Meta-Analysis Based on GWAS Results. Medicine (Baltimore) 2015;94:e1884. doi: 10.1097/MD.0000000000001884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S, et al. Association of single nucleotide polymorphisms in the 3′UTR of ERAP1 gene with essential hypertension in the Northeastern Han Chinese. Gene. 2015;560:211–216. doi: 10.1016/j.gene.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int J Cancer. 2010;127:589–597. doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- 44.Birnbaum S, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 45.Yu Y, et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letra A, Menezes R, Granjeiro JM, Vieira AR. Defining subphenotypes for oral clefts based on dental development. Journal of dental research. 2007;86:986–991. doi: 10.1177/154405910708601013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu, L. et al. Clinical Significance of Long Non-Coding RNA CASC8rs10505477 Polymorphism in Lung Cancer Susceptibility, Platinum-Based Chemotherapy Response, and Toxicity. Int J Environ Res Public Health13, 10.3390/ijerph13060545 (2016). [DOI] [PMC free article] [PubMed]
- 48.Peng Z, Liu L, Wei X, Ling J. Expression of Oct-4, SOX-2, and MYC in dental papilla cells and dental follicle cells during in-vivo tooth development and in-vitro co-culture. Eur J Oral Sci. 2014;122:251–258. doi: 10.1111/eos.12141. [DOI] [PubMed] [Google Scholar]
- 49.Posey JE, et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.