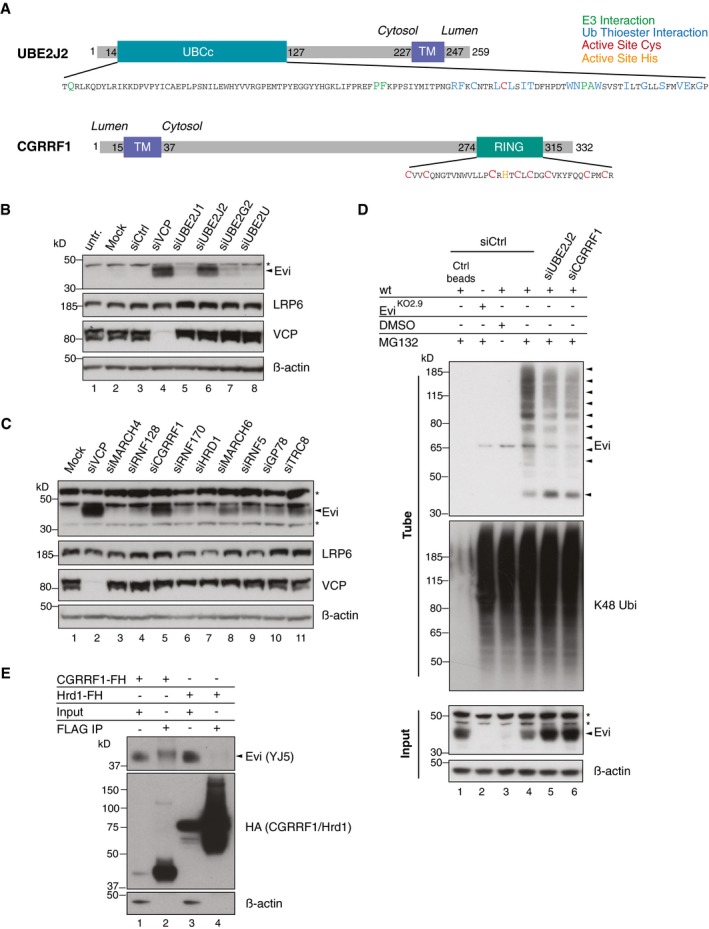
Figure 6. UBE2J2 and CGRRF1 are involved in the degradation and ubiquitination of Evi.
-
ASchematic illustration of UBE2J2 and CGRRF1. The transmembrane (TM) domain of CGRRF1 was predicted using TMHMM server v. 2.0 (Krogh et al, 2001). The core catalytic domain of the E2 conjugating enzyme UBE2J2 (amino acid residues 14–127) is depicted as UBCc. Amino acid residues important for the interaction with E3 ligases are shown in green, residues facing the ubiquitin thioester interaction side in blue, and the active site cysteine in red. The catalytic zinc finger domain of CGRRF1 consists of the typical C3HC4 RING type motif and spans the amino acid residues 274–315. Active site cysteine residues are marked in red and the active site histidine in orange.
-
B, CFollowing reverse transfection of HEK293T cells with pooled siRNA against the indicated proteins, the cell lysates were analyzed by immunoblotting.
-
DFollowing UBE2J2 and CGRRF1 knockdown, HEK293T were treated with 1 μM MG132 for 24 h. Poly‐ubiquitinated proteins were TUBE2 precipitated and stained for endogenous Evi or K48 poly‐ubiquitin. Ctrl agarose beads were used to confirm specificity of the TUBE2 assay for ubiquitinated proteins.
-
ECGRRF1‐FLAG‐HA (FH) and Hrd1‐FLAG‐HA (FH) inducible Flp‐IN HEK293T cells were treated with 1 ng/ml doxycycline for 18 h and 10 μM MG132 for 4 h prior to FLAG IP and subsequent immunoblotting with mouse monoclonal Evi antibody (clone YJ5). All Western blots are representative of three independent experiments. β‐Actin was used as loading control. Specific Evi bands are indicated by arrows and unspecific bands by asterisks.
Source data are available online for this figure.
