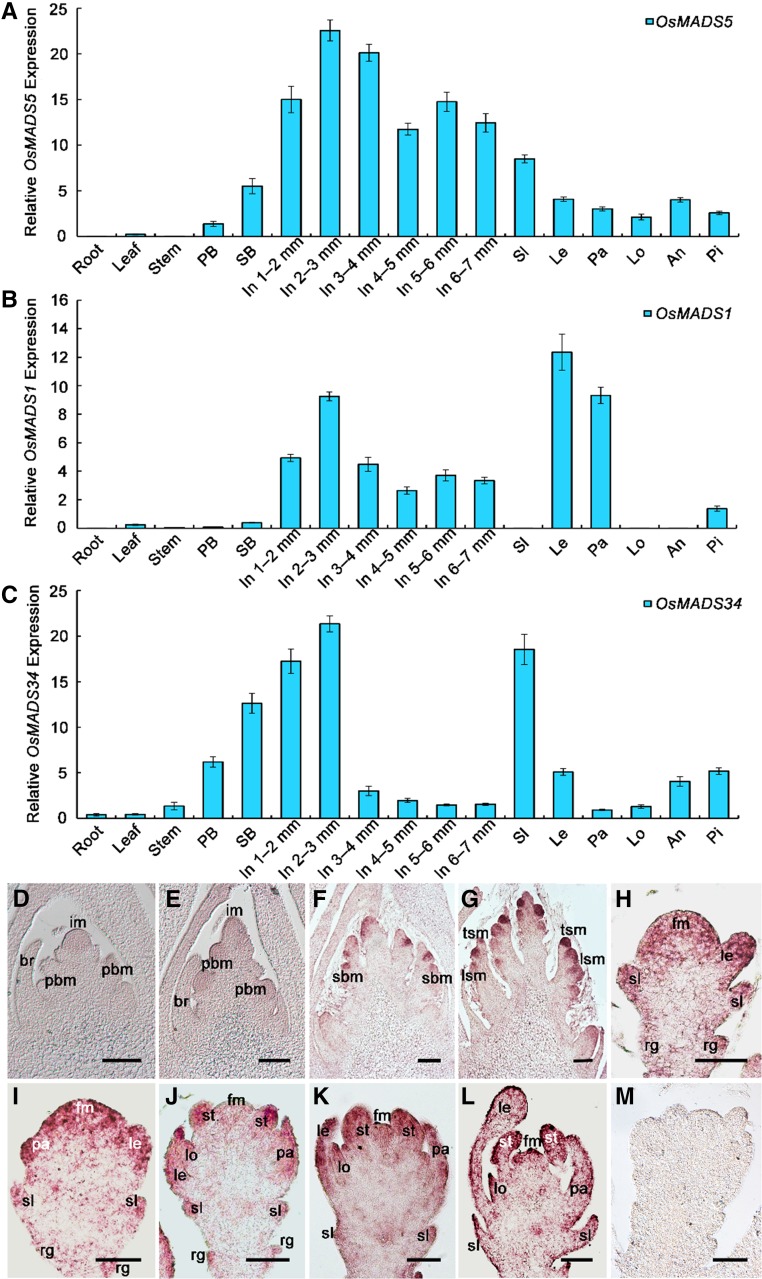
Figure 1.
Expression pattern analysis of rice LOFSEP genes. A to C, qRT-PCR detection of OsMADS5 (A), OsMADS1 (B), and OsMADS34 (C) transcripts. Total RNA was isolated from wild-type roots, leaves, stems, sterile lemmas, lemmas, paleas, lodicules, anthers, and pistils at stage In8 to In9 and also from 0.3- to 7-mm inflorescences. The inflorescence length around 0.3- to 0.6-mm covers the stages of In3 and In4 when the primary branches form and elongate, while inflorescence length 0.6 to 0.9 mm corresponds to stage In5 when the primary branch meristem produces secondary branches, but the SM has not yet formed. The results are presented as mean ± sd. Error bars indicate the SD for three biological replications. D to M, In situ hybridization analysis of OsMADS5 expression in the wild type at stage In3 (D), In4 (E), In5 (F), early In6 (G), Sp3 (H), Sp4 (I), Sp6 (J), Sp7 (K), and Sp8 (L), and at stage Sp6 with sense control (M). an, anther; br, bract; fm, floral meristem; im, florescence meristem; In, inflorescence; le, lemma; lo, lodicule; lsm, lateral spikelet meristem; pa, palea; PB, primary branch; pbm, primary branch meristem; pi, pistil; rg, rudimentary glume; SB, secondary branch; sbm, secondary branch meristem; sl, sterile lemma; tsm, terminal spikelet meristem. Bars = 100 µm (D–G) and 50 µm (H–M).