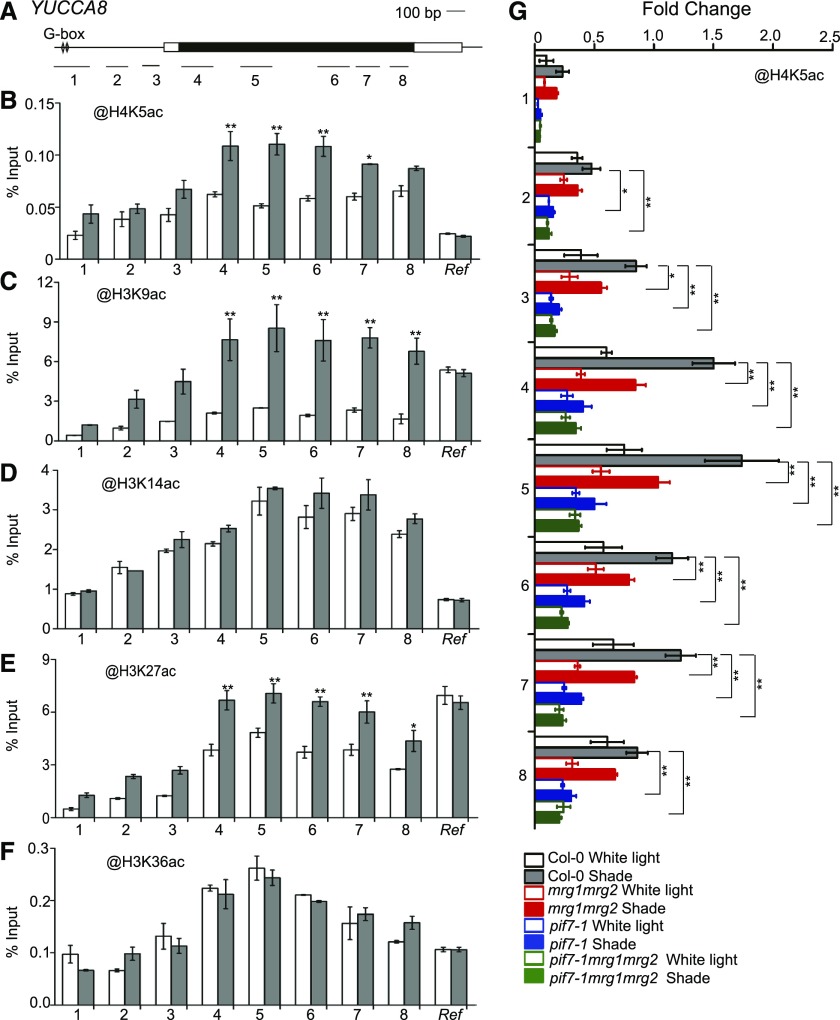
Figure 5.
Histone acetylation modification at YUCCA8 chromatin. A, Schematic of YUCCA8 gene structure. B to F, ChIP-PCR analyses of H4K5ac (B), H3K9ac (C), H3K14ac (D), H3K27ac (E), and H3K36ac (F) at various chromatin regions of YUCCA8 in Col-0 grown under white light and shade conditions. Asterisks indicate where the difference between white light and shade is statistically significant (*P < 0.05, **P < 0.01). G, ChIP-PCR analyses of H4K5ac at YUCCA8 chromatin in Col-0, mrg1mrg2, pif7-1, and pif7-1mrg1mrg2 grown under white light and shade. Asterisks indicate where the difference between wild type and mutant under shade is statistically significant (*P < 0.05, **P < 0.01). Seedlings were grown under white light for 4 d and maintained in white light or transferred to shade for 1 h ChIP and quantitative real-time PCR were performed using specific antibodies and gene primers, as indicated. A region of AT2G39960 served as an internal control and used in normalization. Error bars represent se from three independent biological replicates. Ref, internal control.