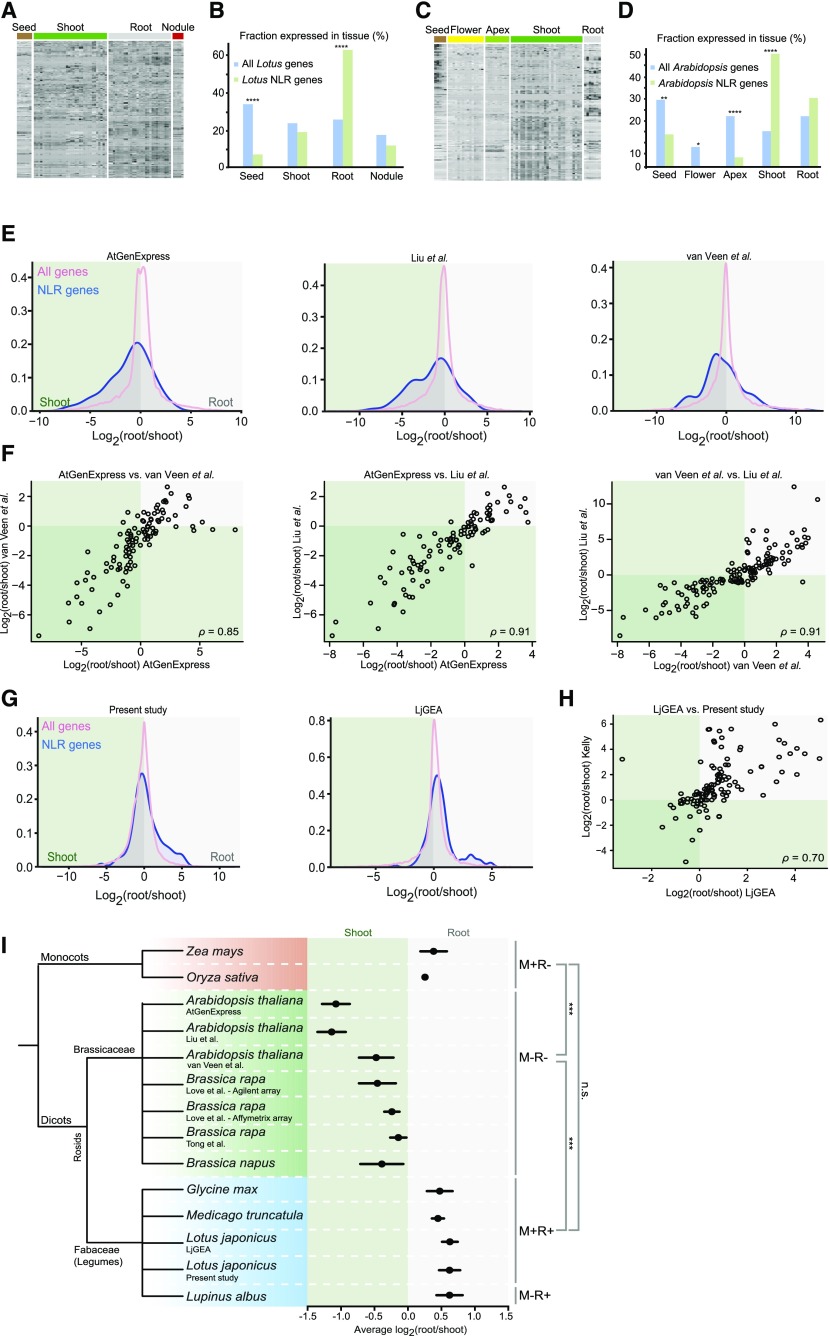
Figure 1.
NLR gene expression patterns across species. A, Expression patterns of 198 putative Lotus NLR genes. B, Enrichment of Lotus NLR genes by tissue type. The fraction of all genes and NLR genes with enriched expression in the given tissue are shown. C, Expression patterns of 160 putative Arabidopsis NLR genes. D, Enrichment of Arabidopsis NLR genes by tissue type. The fraction of all genes and NLR genes with enriched expression in the given tissue are shown. P values indicate the probability that the fraction of NLR genes showing enriched expression in a specific tissue is identical to that of all genes. E, Density plots displaying the distribution of the logarithm of the root/shoot expression ratios of all genes and NLR genes for each Arabidopsis expression data set indicated. See main text for sources. F, Root/shoot expression correlations for Arabidopsis NLR genes from the three data sets shown in Figure 1E. Each circle represents one NLR gene for which expression data are available in both of the datasets compared. G, Density plots displaying the distribution of the logarithm of the root/shoot expression ratios of all genes and NLR genes for each Lotus expression data set indicated. H, Root/shoot expression correlations for Lotus NLR genes between two datasets. Each circle represents one NLR gene for which expression data are available in both datasets. I, Phylogenetic tree of species for which both shoot and root expression data are available, along with their average NLR gene root/shoot expression values (black dots). Error bars indicate SEM. The symbiotic status of each species is indicated on the right. M, mycorrhiza; R, rhizobia; +, engages in endosymbiosis; -, does not engage in endosymbiosis. Significance levels of comparisons between species groups are indicated on the far right. ***P ≤ 0.001. n.s., not significant. Generalized linear model ANOVA was used for calculation of P values. See Supplemental Table S6 for P values for interspecies differences.