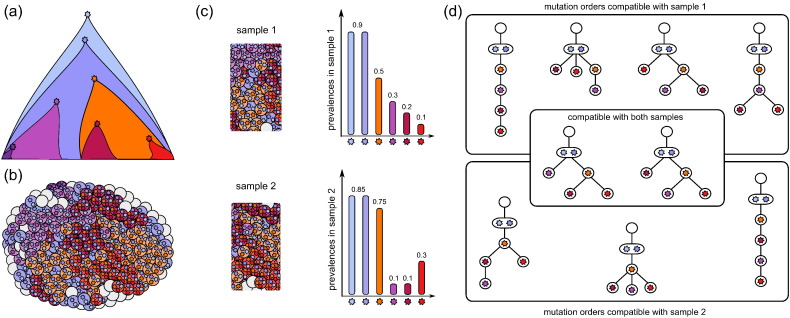
Fig. 1.
(a) Schematic representation of the clonal expansion that shaped the heterogeneous tumour depicted in (b). The colours of the cells represent their belonging to the different subclones. The small stars inside the cells represent the mutations present. (c) Two bulk samples admixed with normal cells (empty grey circles) taken from the tumour in (b). The bar plots depicted next to the samples can be derived from variant allele frequency data obtained by bulk sequencing. Each bar represents the estimated cellular prevalence of one mutation present in the sample. Note that the dark purple mutation on the bottom left of (a) is absent from the frequency plots because it is too low frequency to be detected. (d) Mutation histories compatible with the cell prevalences of sample 1 or sample 2. (Not all compatible trees are depicted.) The two trees in the intersection are compatible with both samples. It can not be inferred from the given data that the left one is the true history that matches the clonal expansion in (a).