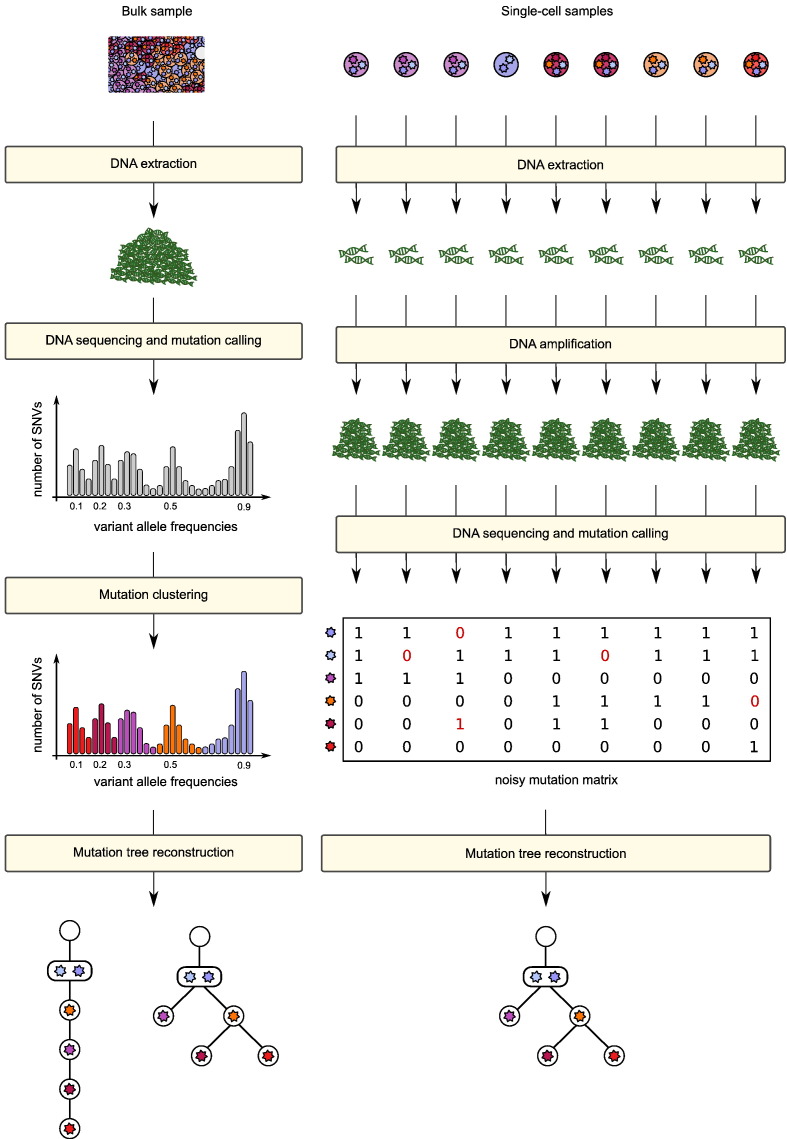
Fig. 3.
Left: Overview of the typical work flow for the reconstruction of mutation histories from bulk tumour samples. DNA is extracted from a bulk sample and sequenced to reveal the admixed mutation profile. Clustering mutations by variant allele frequencies reveals possible subclones and their relative frequency in the admixed sample. Based on this information compatible mutation histories are inferred. Right: Overview of the typical work flow for the reconstruction of mutation histories from single-cell samples. The DNA is extracted from the individual cells and amplified due to the limited starting material. This process does not amplify all genomic sites equally well. The amplified DNA material is then sequenced and mutations are called. The mutation profiles of the individual cells are now combined into a single (noisy) character state matrix that is then used for tree inference.