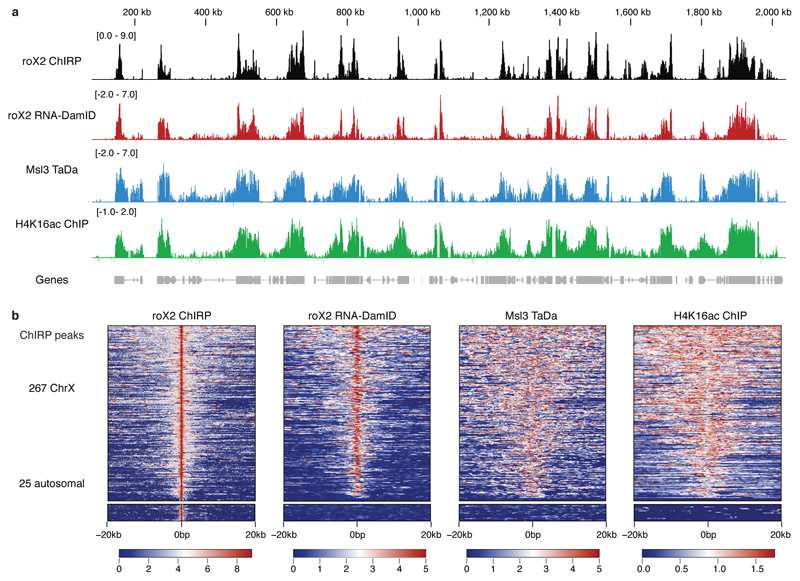
Figure 2. roX2 co-localises with the male-specific lethal complex.
(a) Peaks identified by roX2 RNA-DamID co-localise with roX2 ChIRP 10, Msl3 TaDa and H4K16ac ChIP 11. RNA-DamID scale represents log2 fold change of 3xMS2-roX2 and Dam-MCP compared to 3xMS2 and Dam-MCP (average of two biological replicates). Msl3 TaDa scale is a log2 fold change of Msl3-Dam fusion compared to Dam-alone. Chirp signal is log2 transformed. H4K16ac represents log2 fold change of H4K16ac ChIP over input. (b) Heat map of roX2 RNA-DamID, roX2 ChIRP, Msl3 and H4K16 ChIP signal plotted with over a 20kb window either side of roX2 ChIRP peaks. The majority of ChIRP X chromosome peaks, but not autosomal ChIRP peaks, show an enrichment of RNA-DamID, Msl3 TaDa and H4K16ac ChIP signal.