Abstract
The anterior visual pathway (AVP) conducts visual information from the medulla of the optic lobe via the anterior optic tubercle (AOTU) and bulb (BU) to the ellipsoid body (EB) of the central complex. The anatomically defined neuron classes connecting the AOTU, BU, and EB represent discrete lineages, genetically and developmentally specified sets of cells derived from common progenitors (Omoto et al., 2017). In this paper, we have analyzed the formation of the AVP from early larval to adult stages. The immature fiber tracts of the AVP, formed by secondary neurons of lineages DALcl1/2 and DALv2, assemble into structurally distinct primordia of the AOTU, BU, and EB within the late larval brain. During the early pupal period (P6-P48) these primordia grow in size and differentiate into the definitive subcompartments of the AOTU, BU, and EB. The primordium of the EB has a complex composition. DALv2 neurons form the anterior EB primordium, which starts out as a bilateral structure, then crosses the midline between P6 and P12, and subsequently bends to adopt the ring shape of the mature EB. Columnar neurons of the central complex, generated by the type II lineages DM1-4, form the posterior EB primordium. Starting out as an integral part of the fan-shaped body (FB) primordium, the posterior EB primordium moves forward and merges with the anterior EB primordium. We document the extension of neuropil glia around the nascent EB and BU, and analyze the relationship of primary and secondary neurons of the AVP lineages.
Keywords: Drosophila, brain, lineage, central complex, visual pathway, development, RRID:BDSC_66685, RRID:BDSC_8751, RRID:BDSC_7127, RRID:BDSC_6799, RRID:DGGR_105240, RRID:BDSC_33065, RRID:BDSC_5137, RRID:BDSC_44409, RRID:BDSC_48860, RRID:BDSC_50101, RRID:BDSC_50349, RRID:BDSC_38821, RRID:BDSC_38865, RRID:BDSC_40372, RRID:BDSC_48625
1 Introduction
The central brain of Drosophila is formed by a relatively small number of fixed neural lineages that are produced by genetically unique, stem cell-like neuroblasts (Hartenstein et al., 2008; Ito and Awasaki, 2008). Neural lineages represent genetic modules, as well as structural modules. In the embryo, each neuroblast is defined by the dynamic expression of a specific set of transcription factors. The genetic address provided by these factors plays an essential role in shaping the morphology and function of the corresponding lineage (Brody and Odenwald, 2005; Kohwi and Doe, 2013). Lineages also form structural modules, in that neurons of a lineage form compact clusters of cells and emit axons that bundle into one or two coherent fascicles, the primary and secondary lineage axon tracts. Furthermore, arborizations of a given lineage are spatially confined to one or a few individual neuropil compartment(s). Visualizing lineages by clonal analysis has provided a map of the “macrocircuitry” of the Drosophila brain (Ito et al., 2013; Wong et al., 2013; Yu et al., 2013).
In a recent study we had shown that the neural circuit providing visual input to the central complex, the anterior visual pathway (AVP), is formed by three lineages, DALv2, DALcl1, and DALcl2 (Omoto et al., 2017). Larvally-born (secondary) neurons of DALv2 are confined to the ellipsoid body and its input compartment, the bulb (BU; Larsen et al., 2009; Spindler and Hartenstein, 2011; Wong et al., 2013; Fig. 1). They form the so-called ring (R) neurons of the ellipsoid body, which play a pivotal role in visual memory and complex visually guided behaviors, as well as multiple other functions, controlled by the central complex (Strauss, 2002; Urizar et al., 2007; Neuser et al., 2008; Ofstad et al., 2011; Thran et al., 2013; Martin-Pena et al., 2014; Seelig and Jayaraman, 2015). Different R-neuron classes of the ellipsoid body are defined by their central-peripheral position in the ellipsoid body (Renn et al., 1999; Young and Armstrong, 2010), which in turn is correlated to dendritic location in the bulb. Most easily observed in the horizontal plane of the EB, five discrete domains of the ellipsoid body (EBa/oc/ic/op/ip) can be delineated based on the density of anti-DN-cadherin staining, a global marker of neuropil (Omoto et al., 2017). Ring neurons typically respect boundaries of these domains, generating a basis by which they can be more definitively classified. Most outer ring neurons with axons confined to the outer central (Roc) and anterior domain (Ra) of the EB have dendritic endings in the superior bulb. Some outer central ring neurons innervate the anterior bulb. The inner ring neurons that arborize in the inner central and inner posterior EB domain (Ric, Rip) possess dendrites in the inferior bulb (Omoto et al., 2017; Fig. 1).
Figure 1.
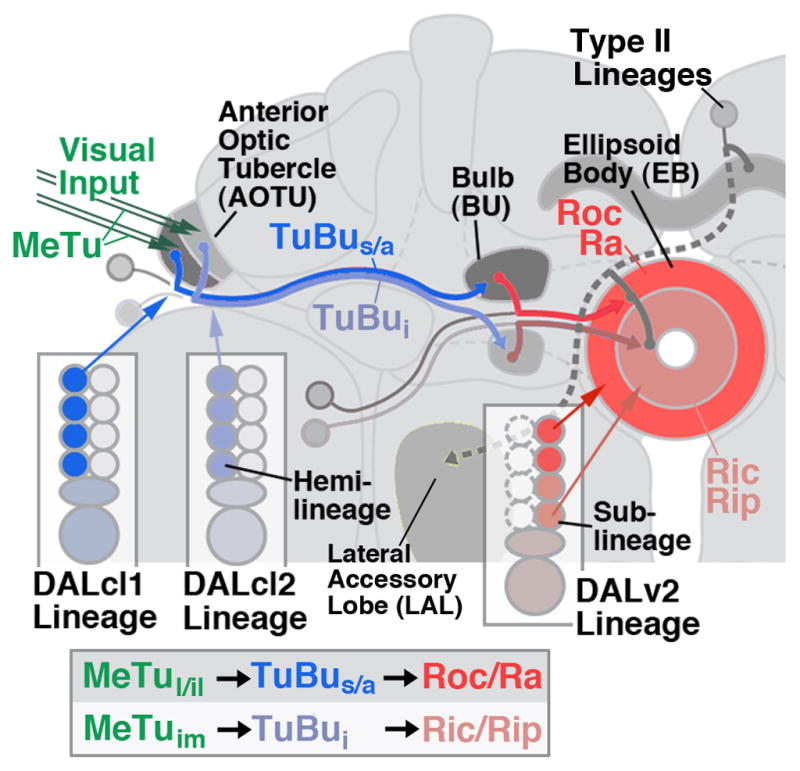
Anterior visual pathway of the Drosophila brain. Schematic frontal view of brain hemisphere depicting the neuronal elements forming the anterior visual pathway, as described in Omoto et al. (2017). Medullo-tubercular (MeTu) neurons project via the anterior optic tract to the lateral and intermediate domain of the anterior optic tubercle; AOTU). Two hemilineages, DALcl1d and DALcl2d, form the tuberculo-bulbar (TuBu) neurons that connect the tubercle to the bulb (BU). Lineage DALv2, formed by one surviving hemilineage (Kumar et al., 2009), forms multiple classes of ring (R) neurons with dendrites in the bulb, and axons in the ellipsoid body (EB). These classes (symbolized by dark red and light red shading) comprise sublineages born at different developmental intervals (J.L. and V.H., unpublished). Box at bottom of diagram presents linear arrangement of the elements of the AVP.
Input to the bulb (BU) originates in the anterior optic tubercle (AOTU), which in turn is a recipient of visual interneurons from the medulla. Two hemilineages (sets of neurons derived from the unequal division of the ganglion mother cells; Truman et al., 2010), DALcl1d and DALcl2d (Omoto et al., 2017), form a topographically ordered projection from AOTU to BU that respects the boundaries set by the subclasses of DALv2 ring neurons. DALcl1 and DALcl2 are two neighboring lineages with cell bodies in the dorso-anterior cortex. Both include a hemilineage whose axon tract passes dorsal of the peduncle (DALcl1/DALcl2d), and another one passing ventrally (DALcl1v/DALcl2v; Lovick et al., 2013). Only the dorsal hemilineages contribute to the AVP. DALcl1d neurons connect the lateral domain of the AOTU (AOTUl, AOTUil) with the superior and anterior BU (TuBus; TUBua; Fig. 1). DALcl2d neurons connect the intermediate medial domain of the AOTU (AOTUi,) to the inferior BU (TuBui; Fig. 1). The AOTUl and AOTUi receive input from the medulla of the optic lobe via different populations of multi-columnar medullary projection neurons (MeTu). Functional studies of DALcl1/2 input to the bulb demonstrated that the spatially and developmentally discrete subpopulations of neurons targeting the superior bulb have properties that fundamentally differ from the inferior bulb pathway: the former is activated by small ipsilateral stimuli and projects in a precise retinotopic manner onto the bulb; the latter are inhibited by ipsilateral stimuli, and are activated by contralateral stimuli distributed widely over the visual field (Omoto et al., 2017).
Similar to the AVP itself, output pathways from the ellipsoid body are also structured around lineages. For example, PB-EB-gall neurons (Wolff et al., 2015) are sublineages of four large (type II) lineages (Yang et al., 2013). These include DM1/DPMm1, DM2/DPMpm1, DM3/DPMpm2, DM4/CM4 (Bello et al., 2008; Boone and Doe, 2008; Bowmann et al., 2008; Ito et al., 2013; Wong et al., 2013; Yu et al., 2013), from here on onward called DM1-DM4 in the text. PB-EB-gall neurons form a topographically highly ordered pathway that interconnects small segments of the protocerebral bridge (PB; the posterior-most compartment of the central complex) with the ellipsoid body and the lateral accessory lobe, a known premotor area of the insect brain (Namiki and Kanzaki, 2016).
With the properties summarized above, the AVP exemplifies a circuit where structurally and functionally discrete classes of neurons represent developmental units, that is, components of a small number of lineages. In the present paper, we have investigated the development of the AVP. Previous works had already shed light on developmental and evolutionary aspects of the central complex (Renn, 1999; Loesel et al., 2002; Young and Armstrong, 2010; Riebli et al., 2013); however, a detailed analysis of the sequential steps leading up to the formation of the ellipsoid body and its input pathway had not been carried out. In this work, using markers expressed throughout development, we were able to follow the first appearance of the different elements of the AVP in the larva, and map the larval primordia of this pathway. Like larval primordia of adult neuropil compartments in general, primordia of the AVP are formed by the undifferentiated axon tracts of lineages DALv2, DALcl1/2, and the posterior type II lineages, which represent dense bundles with filopodia, lacking the expression of synaptic markers that are characteristic for differentiated neuropil (Omoto et al., 2015). During the first 48 hours of metamorphosis, these primordia grow in size and differentiate into the definitive subcompartments of the AOTU, BU, and EB. Of particular interest was the formation of the EB, which consists of two very different populations of neurons: wide-field R neurons of lineage DALv2, located in the anterior brain, and small-field (columnar) neurons of the DM1-4 lineages, located in the posterior brain. We can show that these two groups initially form two separate primordia which later merge into the EB. Finally, we provide a first description of the projection of the primary neurons of lineages DALv2 and DALcl1/2. Given that a central complex and AVP, in the anatomical sense, are not yet formed in the larval brain, we ask what compartments are innervated by the primary neurons of these lineages, and how they relate to the adult central complex. Our data provide a foundation for future studies addressing the genetic and developmental mechanisms by which complex, homotopically ordered pathways are controlled.
2 Materials and Methods
2.1. Fly Lines
insc-Gal4 (Mz1407; Betschinger et al, 2006; Bloomington Drosophila Stock Center (BDSC) #8751; RRID:BDSC_8751), poxn-Gal4 (Boll and Noll, 2002; kindly provided by Dr. H. Reichert; RRID:BDSC_66685), per-Gal4 (Kaneko and Hall, 2000; #7127, BDSC; RRID:BDSC_7127); NP6520-Gal4 (Awasaki et al., 2008) (Kyoto DrosophilaGeneticResourceCenter; RRID:DGGR_105240), Nrv2-Gal4 (Sun et al., 1999; kindly provided by Dr. P. Salvaterra; RRID:BDSC_6799); UAS-DenMark::mCherry, UAS-Syt::GFP (Nicolai et al., 2010; #33065; RRID:BDSC_33065); UAS-mcd8::GFP (Lee et al., 1999; #5137, BDSC; RRID:BDSC_5137); 442-Gal4 (Hitier et al., 2000; Chen and Hing, 2008; kindly provided by Dr.H.Hing); EB1-Gal4 (Renn et al., 1999; # 44409; RRID:BDSC_44409); R19G02-Gal4, R40G10-Gal4, R48B06-Gal4, R52B11-Gal4, R53B08-Gal4, R83H08-Gal4, R82E10-Gal4 (Janelia Farm GAL4 Stock Collection, Jenett et al., 2012; #48860 (RRID:BDSC_48860), #50101 (RRID:BDSC_50101), #50349 (RRID:BDSC_50349), #38821 (RRID:BDSC_38821), #38865 (RRID:BDSC_38865), #40372 (RRID:BDSC_40372), #48625 (RRID:BDSC_48625)).
Flies were grown at 25°C using standard fly media unless otherwise noted.
2.2. Immunohistochemistry
The following primary antibodies were used: mouse anti-Neurotactin (Nrt, BP106; RRID:AB_528404), mouse anti-Neuroglian (Nrg, BP104; RRID:AB_528402), rat anti-DN-cadherin (DN-Ex #8; RRID:AB_2314331), mouse anti-Bruchpilot (Nc82; RRID:AB_2314868). All antibodies from Developmental Studies Hybridoma Bank (DSHB, University of Iowa, Iowa City, Iowa; each diluted 1:10). For fluorescent staining, the following secondary antibodies were used: Alexa Fluor 546 goat anti-Mouse IgG (H+L) (#A11030; Invitrogen, Carlsbad, CA; used at 1:500) and Cy5 goat anti-Rat IgG (H+L) (112-175-143; Jackson Immunoresearch, West Grove, PA; used at 1:400).
All larvae and adults were grown at 25°C on standard food media. Adults were aged 3 to 5 days post-eclosion before dissection. For antibody labeling, standard procedures were followed (Ashburner, 1989). Briefly, dissected brains were fixed with phosphate buffered saline (PBS), pH 7.4, containing 4% paraformaldehyde for 25 – 30 min. They were then washed with 1× PBS, pH 7.4, containing 0.1 % Triton X-100 for 3 X 10 min. Samples were then incubated in blocking buffer (2% bovine serum albumin (BSA) in 1X PBS, pH 7.4, containing 0.1 % Triton X-100) for 1 hour at room temperature. They were then incubated with primary antibody diluted in blocking buffer overnight at 4°C. They were subsequently washed 3 X 15 min in 1X PBS, pH 7.4, containing 0.1 % Triton X-100 at room temperature, followed by blocking buffer for 20 min. Samples were incubated with secondary antibody diluted in blocking buffer overnight at 4°C, followed by washes in 1X PBS, pH 7.4, containing 0.1 % Triton X-100 for 3 X 15 min, and mounting in Vectashield (Vector Laboratories).
2.3. Markers
The DN-cadherin antibody (DSHB DN-EX #8), a marker for neuropil, is a mouse monoclonal antibody raised against a peptide encoded by exon 8, amino acid residues 1210–1272 of the Drosophila CadN gene. The antibody detected two major bands, 300 kDa and 200 kDa molecular weights on Western blot of S2 cells only after transfection with a cDNA encoding the DN-cadherin protein (Iwai et al., 1997). In addition, the specificity of this antibody was tested with immunostaining of Drosophila embryos. Signal was hardly detectable in homozygous mutant, l(2)36DaM19 with nonsense mutation causes premature termination of protein translation. In contrast, this antibody gave a signal in mutant embryos with N-cadherin transgene.
The Neurotactin antibody (DSHB BP106) labels secondary neurons and their axons. It is a mouse monoclonal antibody raised against the first 280 aminoterminal amino acid residues (Hortsch et al., 1990) of the Drosophila Nrt gene. The monoclonal antibody detected the same Drosophila embryonic pattern to that of a polyclonal antisera raised against a fusion protein using part of the Neurotactin cDNA (Hortsch et al., 1990). In addition, another monoclonal antibody, MAb E1C, against Neurotactin gave a similar expression pattern in Drosophila embryos to that of BP106 (Piovant and Léna, 1988).
The Neuroglian antibody (DSHB BP104) labels secondary neurons and axons in the adult brain. It is a mouse monoclonal antibody from a library generated against isolated Drosophila embryonic nerve cords (Bieber et al., 1989). The Neuroglian antibody was used to purify protein from whole embryo extracts by immunoaffinity chromatography. Protein microsequencing of the purified protein was performed to determine that the 18 N-terminal amino acids that is identical to the sequence determined for the N-terminus of the protein based on a full-length cDNA clone (Bieber et al., 1989).
Monoclonal mouse anti-Bruchpilot antibody (DSHB Nc82) labels synapses. Antigen: Raised against adult Drosophila head homogenates. The specific immunogen was identified as Bruchpilot (Wagh et al., 2006). In Western blots of homogenized Drosophila heads, the antibody specifically recognized two proteins of 190 and 170 kDa apparent size which were later found to be part of the same transcription unit of the bruchpilot gene. In vivo, the antibody recognizes brain neuropil as well as synaptic active zones during most stages of Drosophila brain development.
2.4. Confocal Microscopy
Staged Drosophila larval and adult brains labeled with suitable markers were viewed as whole-mounts by confocal microscopy [LSM 700 Imager M2 using Zen 2009 (Carl Zeiss Inc.); lenses: 40× oil (numerical aperture 1.3)]. Complete series of optical sections were taken at 2-μm intervals. Captured images were processed by ImageJ or FIJI (National Institutes of Health, http://rsbweb.nih.gov/ij/ and http://fiji.sc/) and Adobe Photoshop.
3 Results
3.1. List of Abbreviations used throughout the Figures
ACO antennal lobe commissure; ADC anterior dorsal commissure; AOT anterior optic tract; CA calyx; DALcl1, DALcl2, DALv2 dorso-anterior lateral type I lineages; DM1/DPMm1, DM2/DPMpm1, DM3/DPMpm2, DM4/CM4 dorso-posterior medial type II lineages; FBapl anterior plexus of fan-shaped body; FBppl posterior plexus of fan-shaped body; FrMC fronto-medial commissure; GC great commissure; IPa anterior domain of inferior protocerebrum; LAL lateral accessory lobe; LEa anterior lateral ellipsoid body fascicle; LEp posterior lateral ellipsoid body fascicle; MB mushroom body; MDBchi chiasm of median bundle; ML medial lobe; MeTu axon bundle formed by medullo-tubercular neurons; PED peduncle; prAOTU primordium of anterior optic tubercle (il intermediate lateral subdivision of prAOTU; im intermediate-medial subdivision of prAOTU; in intermediate subdivision of prAOTU; l lateral subdivision of prAOTU; la anterior lateral subdivision of prAOTU; lc central lateral subdivision of prAOTU; lp posterior lateral subdivision of prAOTU; m medial subdivision of prAOTU; prBU primordium of bulb; prBUs primordium of superior bulb; prBUi primordium inferior bulb; prEBa primordium of anterior ellipsoid body; prEBp primordium of posterior ellipsoid body; prFB primordium of fan-shaped body; prLALg primordium of gall of lateral accessory complex; prNO primordium of noduli; prPB priordium of protocerebral bridge; R axon bundle formed by ring neurons (lineage DALv2); Ric inner central domain of ellipsoid body; Rip inner posterior domain of ellipsoid body; Roc outer central domain of ellipsoid body; SAC superior arch commissure; SEC supraellipsoid body commissure; SLP superior lateral protocerebrum; SMP superior medial protocerebrum; SuEC subellipsoid body commissure; TuBu axon bundle formed by tuberculo-bulbar neurons (lineages DALcl1/2); TuBua tuberculo-bulbar neurons projecting to anterior bulb; TuBui tuberculo-bulbar neurons projecting to inferior bulb; TuBus tuberculo-bulbar neurons projecting to superior bulb; VL vertical lobe; VLP ventrolateral protocerebrum
3.2. Axonal projections and neuropil primordia of the AVP form in the third instar larva
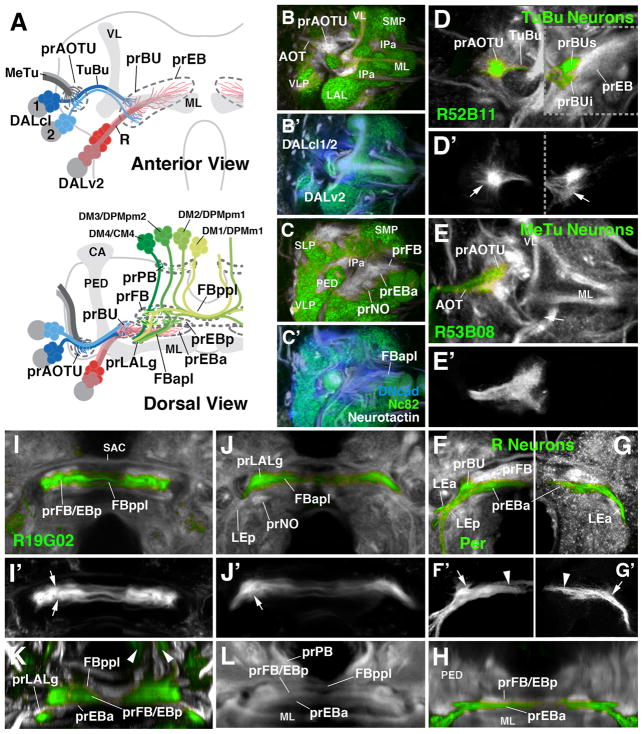
The central elements of the anterior visual pathway, formed by secondary neurons of lineages DALcl1, DALcl2, and DALv2 (Omoto et al., 2017), are born between 30 and ~100h after hatching and extend axons that coalesce in lineage-associated tracts (secondary axon tracts; SATs), but do not arborize and differentiate prior to metamorphosis (Lovick and Hartenstein, 2015; Lovick et al., 2016). In the late L3 larva, the anterior visual pathway appears as discrete axon bundles that interconnect the primordia of the medulla, optic tubercle, bulb, and ellipsoid body, respectively (Fig. 2A). The primordia themselves can be recognized as tufts of filopodia branching off the axon bundles, expressing high levels of DN-cadherin. These filopodial tufts are negative for markers of differentiated synapses, such as Bruchpilot (Brp or Nc82; Wagh et al., 2006), which sets them apart from the surrounding differentiated, Brp-positive larval neuropil (Fig. 2B–C′). We can recognize the primordia of the anterior optic tubercle (prAOTU), the anterior ellipsoid body (prEBa) that is merged with the primordia of the bulb (prBU), the posterior ellipsoid body (prEBp), the fan-shaped body (prFB), protocerebral bridge (prPB) and noduli (prNO), as described in the following and illustrated by Fig. 2.
Figure 2.
Larval primordia of the central complex and anterior visual pathway. (A) Schematic view of larval brain hemisphere (top: anterior view, dorsal up; bottom: dorsal view, posterior up). Secondary lineages contributing to the primordia of anterior visual pathway and central complex are rendered in different colors; mushroom body (gray) is shown as reference. (B, B′, C, C′) Z-projections of frontal confocal sections of late larval brain hemisphere labeled with anti-Bruchpilot (Nc82; green), anti-DN-cadherin (DNcad; white in B, C; blue in B′, C′) and anti-Neurotactin (BP106; white in B′, C′). Note domains which are negative for Nc82, indicating absence of synapses; these domains are positive for DNcad and BP106, showing presence of (undifferentiated) secondary fibers and filopodia which make up primordia of adult-specific compartments. Labeled are primordia of anterior optic tubercle (prAOTU), anterior ellipsoid body (prEBa), bulb (prBU), fan-shaped body (prFB), noduli (prNO. (D–K): Z-projections of confocal sections of late larval brain hemispheres in which individual cell classes forming part of anterior visual pathway and central complex are labeled by specific Gal4 drivers (green). In (D–L), DNcad antibody labels neuropil (white); (D′–G′) show only GFP expression. (D–G′) and (I, J) show frontal sections, (H) and (K, L) are digitally tilted dorsal views. (D, D′) Tuberculo-bulbar (TuBu) neurons of DALcl1/2 lineages, labeled by R52B11-Gal4. Shown to the left of hatched line is a more anterior plane, containing tufts of proximal filopodia (marked by arrow in D′) that constitute the primordium of the anterior optic tubercle. To the right of the hatched line is a slightly more posterior plane, featuring endings and distal filopodia of TuBu neurons, which form the primordia of the superior and inferior bulb (prBUs, prBUi), respectively. (E, E′) Axonal endings of medullo-tubercular (MeTu) neurons in the primordium of the anterior optic tubercle, labeled by R53B08-Gal4. (F–H) Ring (R) neurons of DALv2 lineage, labeled by Period (Per)-Gal4 (F, F′, H) or as MARCM clone (G, G′). DALv2 neurons form a straight, medially-directed bundle, the anterior lateral ellipsoid body tract (LEa), which extends posterior to the medial lobe of mushroom body (ML in panel A) to end close to the midline. Filopodial tufts of these axons form the primordia of the anterior ellipsoid body (prEBa; arrowheads in F′, G′) and, further laterally, the bulb (prBU; arrows in F′, G′). (I–L) Subset of columnar neurons of the central complex, labeled by R19G02-Gal4. These neurons form part of four type II lineages, DM1/DPMm1, DM2/DPMpm1, DM3/DPMpm2, DM4/CM4, as shown in panel (A). Axons project forward and proximal filopodia form the primordium of the protocerebral bridge (arrowheads in K; prPB in A). Axons then branch and turn medially, forming the posterior plexus of the fan-shaped body (FBppl in A, I, K). Filopodial tufts branch in the primordium of the fan-shaped body (A, I, K) and the posterior ellipsoid body (prFB/EBp in I, K). Axons continue forward, branching towards medially and laterally, forming the anterior plexus of the fan-shaped body (FBapl; A, J). Distal tips of this projection defines the primordium of the gall of the LAL (prLALg; A, J, K). For other abbreviations, see List of Abbreviations. Bar: 25μm
The larval primordium of the anterior optic tubercle (prAOTU; labeled by R52B11-Gal4 in Fig. 2D, D′) forms a hemispherical domain at the anterior neuropil surface, laterally adjacent to the vertical lobe of the mushroom body. Filopodia foreshadowing the dendrites of tuberculo-bulbar (TuBu) neurons, formed by DALcl1d and DALcl2d, diffusely overlap in this domain with axonal filopodia of the medullary MeTu neurons from the optic lobe, labeled by R53B08-Gal4 (Fig. 2E, E′). The TuBu tract projects medially across the peduncle of the mushroom body and terminates in two separate filopodial tufts which pioneer the superior and inferior bulb, respectively (prBUs, prBUi; Fig. 2D). The bulb primordia are located in the anterior inferior protocerebrum (IPa) compartment, posteriorly adjacent to the medial lobe of the mushroom body (Fig. 2A).
Aside from the endings of DALcl1/2, the DALv2 lineage (labeled by period-Gal4 or poxn-Gal4; Kaneko and Hall, 2000; Boll and Noll, 2002; Larsen et al., 2009) contributes to the primordium of the bulb and, in addition, pioneers the ellipsoid body. The DALv2 tract forms a straight, postero-medially directed bundle that passes underneath the medial lobe of the mushroom body before turning medially and terminating well short of the midline (Fig. 2A, F, G). This bundle is called the anterior lateral ellipsoid fascicle (LEa; Strausfeld, 1976; Lovick et al., 2013; Wong et al., 2013 Fig. 2F). Proximal filopodia branching off of DALv2 axons intermingle with the DALcl1/2 filopodia in the bulb primordium; more distal filopodia of DALv2 constitute the main, anterior part of the primordium of the ellipsoid body (prEBa; Fig. 2A, F–H). An anatomical distinction between different classes of R neurons is not yet apparent within the prEBa, or the prBU. The tract of BAmv1 neurons, which also expresses per-Gal4 (Larsen et al., 2009), converges onto the anterior prEBa from ventrally. It forms the posterior lateral ellipsoid fascicle (LEp; Lovick et al., 2013; Wong et al., 2013; Fig. 2F).
In the adult EB, DALv2 ring neurons and columnar neurons produced by the type II lineages DM1-4, overlap widely (Omoto et al., 2017). By contrast, in the L3 larva, both populations of cells form discrete, spatially separate primordia, as shown by the marker R19G02-Gal4 which labels specific subsets of columnar neurons, notably the ones connecting the protocerebral bridge, ellipsoid body, and LALgall (PB-EB-gall neurons; Wolff et al., 2015). Filopodial tufts of these neurons form the posterior component of the ellipsoid body primordium (prEBp) in the late larval brain (Fig. 2A, I–L). Interestingly, this structure arises as a part of the previously defined fan-shaped body primordium (prFB; Pereanu et al., 2010; Riebli et al., 2013). Axon tracts DM1-4 project anteriorly, passing the primordium of the protocerebral bridge (prPB; Fig. 2A bottom panel, K, L), and then turn medially to form a dense plexus of fibers at the posterior boundary of the prFB (Fig. 2A, I, I′). This posterior plexus of the fan-shaped body (FBppl) remains visible into the adult stage as a distinctive anatomical landmark of the central complex. Further anteriorly, the combined fan-shaped body/posterior ellipsoid body primordium (prFB/EBp) is formed by dense, anteriorly directed filopodial tufts emerging from the plexus (Fig. 2A, I, I′). The prFB/EBp directly abuts the anterior prEBa, formed by the DALv2 tract/filopodia (Fig. 2A, K, L). As described in the following section, the prEBp separates from the prFB at the onset of metamorphosis, moving anteriorly and merging with the prEBa.
A subset of R19G02-Gal4-positive fibers continue anteriorly, then turn medially or laterally, forming a second plexus at the anterior boundary of the fan-shaped body primordium (Fig. 2A, J, J′). This anterior plexus of the fan-shaped body (FBapl) also develops into a prominent fiber system of the adult brain, capping the fan-shaped body anteriorly, and the ellipsoid body dorsally (Omoto et al., 2017). The R19G02-Gal4-positive elements of the FBapl terminate in a small focus at the dorso-lateral surface of the lateral accessory lobe (Fig. 2A, J–K), pioneering the gall of the lateral accessory lobe (prLALg), which receives axonal terminals of the PB-EB-gall neurons of the adult brain.
3.3. Morphogenesis of the anterior ellipsoid body and bulb
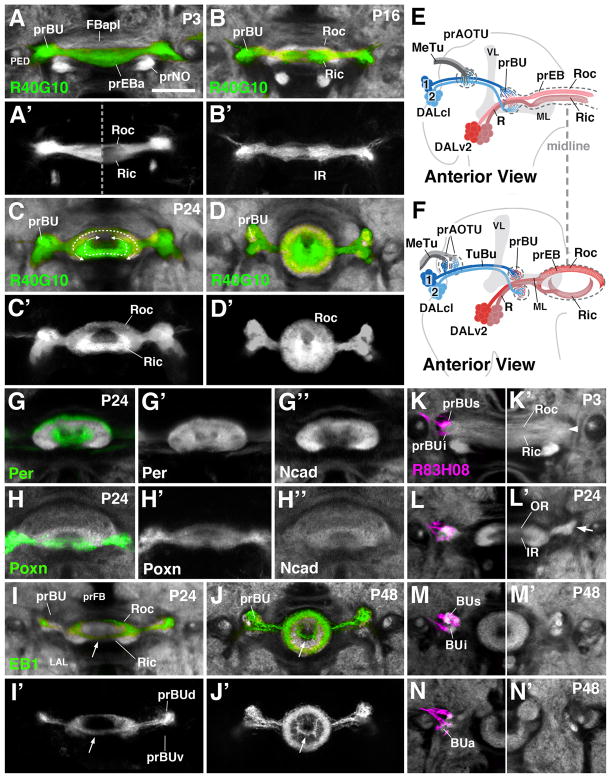
During the early pupal period (P6-P48), the primordia of the anterior visual pathway grow in size, and the ellipsoid body takes on its definitive, layered shape. During the first 12h after onset of metamorphosis, the axons of DALv2 derived R-neurons have crossed the brain midline, and sorted out into a dorsal component, the future outer ring (Roc) and a ventral component, which foreshadows the inner ring (Ric; Fig. 3A, A′, B, B′, E). These two layers, which form the anterior primordium of the ellipsoid body (prEBa), are recognizable on the basis of differential labeling with Gal4 driver lines specific for R-neuron subsets, as well as anti-DN-cadherin. Particularly helpful is the driver R40G10, expressed from larval to adult stages at high levels in the inner ring, and weaker in the outer ring (Fig. 3A–D′). Between P12 and P48, R axons bend, with the result that the initially straight Roc and Ric domains adopt their characteristic ring shape. Roc fibers bend ventrally, around the inner mass of Ric fibers, whereas the Ric fibers grow dorsally, as seen most clearly at around P24 using labeling with several different drivers, as well as the global neuropil marker DN-cadherin (Fig. 3C, C′, F, G–H″). Inner and outer EB neuropils have completed their ring shape prior to P48 (Fig. 3D, D′). The morphogenesis of some R-neuron populations deviates from this general pattern. In particular, some populations of outer ring neurons (e.g., R2 neurons expressing EB1; Fig. 3I–J′) form a pattern that describes a complete ring structure already at P24 (Fig. 3I, I′). It is possible that in this case, the ventral arch (arrow in Fig. 3I, I′) is formed by the R2 axons that enter the EB from anteriorly near its center (arrow in Fig. 3J, J′) and spread outward from there.
Figure 3.
Morphogenesis of the ellipsoid body and bulb. (A–D′) and (G–N′) Z-projections of frontal confocal sections of pupal brain hemispheres at different stages, indicated in upper left corner of panels (P3 3hrs after puparium formation (APF); P16 16hrs APF; P24 24hrs APF; P48 48hrs APF). In panels (A–D) and (G–N), specific neuron populations are labeled by Gal4-drivers indicated at lower left corner of panels; antibody against DN-cadherin marks neuropil (white). (A–D′) R40G10-Gal4 labels preferentially inner ring (Ric) neurons throughout development. (E, F) Schematic anterior view of pupal brain hemisphere at 6hrs (E) and 24hrs (F) after puparium formation (APF). Secondary lineages contributing to the primordia of the anterior visual pathway are rendered in different colors; mushroom body (gray) is shown as reference. (G–H″) Per-Gal4 and Poxn-Gal4 are expressed in all R-neurons, but allow one to distinguish the inner from outer ring during early pupal stages. (I–J′) EB1-Gal labels R2 class of outer ring neurons. (K–N): R83H08-Gal4 (magenta) is expressed in tuberculo-bulbar neurons and labels the developing bulb primordium (prBUs, prBUi, prBUa) of left brain hemisphere. Panels (K′–N′) show opposite hemisphere, labeled only with anti-DN-cadherin (white). For other abbreviations, see List of Abbreviations. Bar: 25μm
The primordium of the bulb (prBU) is recognizable as a DNcad-rich domain separable from the ellipsoid body primordium from stage P6 onward. Flanking the prEB laterally, the superior part of the prBU is located at the level of the presumptive outer ring, the inferior part next to the presumptive inner ring neurons (Fig. 3A, B, E, K). Labeling of DALcl1/2d neurons at this stage with R83H08-Gal4 shows a clear separation of dorsal and ventral axon terminals (Fig. 3K), demarcating the primordia of the BUs and BUi, respectively. However, based on neuropil markers (anti-DN-cadherin), a boundary between presumptive BUs and BUi is not apparent yet (arrowhead in Fig. 3K′). This boundary appears at around P24, when the bending of DALv2 axons into a ring-shape is well under way and, as a result, the primordium of the ellipsoid body has become narrower and spatially separated from the bulb primordium (Fig. 3L, L′). A lateral indentation demarcates the boundary between BUs and BUi (arrow in Fig. 3L′). At this stage, dendritic branches of inner and outer DALv2 R-neurons have also separated; as shown in Fig. 3H, dendrites of outer ring neurons, labeled by EB1-Gal4, are restricted to the dorsal bulb. Morphogenesis of the bulb is complete by P48, when neuropil markers delineate discrete BUs, BUi, and BUa (Fig. 3M–N′).
3.4. Differentiation of the anterior optic tubercle
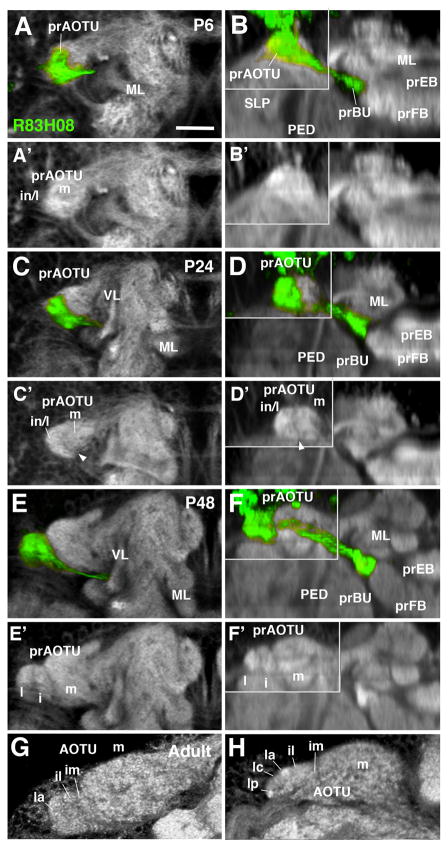
The AOTU of the adult brain is comprised of a medial division (AOTUm), which receives input from the lobula, and an intermediate and lateral subdivision (AOTUi, AOTUl) targeted by medullary MeTu neurons, and connected to the bulb by DALcl1/2d neurons. The AOTUl is further subdivided into three smaller domains, the anterior, intermediate, and posterior AOTUl (Omoto et al., 2017). The gradual differentiation of the AOTU from its larval primordium can be followed by labeling DALcl1/2d neurons with R83H08-Gal4, which is expressed throughout larval and pupal development (Fig. 4). As described above, the larval prAOTU is represented by a small, homogenous domain in which the distal filopodia of afferent optic lobe interneurons and proximal filopodia of DALcl1/2d neurons overlap completely. During the first 12h of metamorphosis the prAOTU grows significantly, and differentiates into a ventro-lateral, DN-cadherin-rich domain which is innervated by the bulk of R83H08-Gal4-positive fibers, and a dorso-medial, DN-cadherin-poor region (Fig. 4A–B′). The former domain represents the primordium of the intermediate and lateral tubercle (prAOTUi/l), and the latter one the primordium of the medial tubercle (prAOTUm), which does not receive DALcl1/2d dendrites. By P24, the axis of the prAOTU has tilted more towards the horizontal, bringing the prAOTUi/l to a position laterally adjacent to the prAOTUm, at which it remains until the adult stage (Fig. 4C–D′). However, the subdivision of the lateral part of the tubercle into AOTUim, AOTUil, and AOTUla-p is not yet evident at this stage. This subdivision starts to be visible at around P48, when the lateral tubercle clearly shows two separate domains, set apart by a sharp boundary (Fig. 4E–F′). The lateral domain (AOTUl) is already characterized by a higher intensity of DNcad expression than the intermediate domain (AOTUi; Fig. 4E′, F′). The further distinction of the three subdomains of the AOTUl, arranged along the anterior-posterior (AOTUla, AOTUi, AOTUp) becomes morphologically visible around the time of eclosion (Fig. 4G, H).
Figure 4.
Morphogenesis of the anterior optic tubercle. Panels show Z-projections of frontal confocal sections (left column; A, C, E, G) and corresponding, digitally-tilted horizontal sections (right column; B, D, F, H) of brain hemisphere at different stages of development, from 6hrs APF (P6) to Adult (stages indicated in upper right corner of panels). In panels of right column, the area in upper left boxed by gray line represents a dorsal level, including the prAOTU; remainder of the panels show horizontal sections at a more ventral level, including the prBU. Proximal arborizations of TuBu neurons, labeled by R83H08-Gal4 (green), mark the developing primordium of the anterior optic tubercle (prAOTU); distal arborizations outline the primordium of the bulb (prBU). Neuropil is labeled by anti-DN-cadherin (white). In early pupa (P6; A–B′) the prAOTU forms an undifferentiated, ovoid, DN-cadherin-rich domain at the anterior-lateral surface of the superior lateral protocerebrum (SLP). GFP-positive fibers of TuBu neurons are largely restricted to the lateral half of the prAOTU (A), which will give rise to the intermediate and lateral subdomains of this compartment (in/l in A′). The medial part of the prAOTU (m in A′) does not receive GFP-positive fibers. At P24 (C–D′), a sharp boundary separates the medial from the intermediate/lateral subdomain (arrowhead in C′, D′). At P48 (E–F′), the GFP-positive domain has differentiated into a lateral (l) and intermediate (i) subdomain. During late pupal development, these become further subdivided into smaller subdomains, including the intermediate medial (im), intermediate lateral (il), lateral anterior (la), lateral central (lc), and lateral posterior (lp; G, H). For other abbreviations: see List of Abbreviations. Bar: 25μm.
3.5. Formation of the ellipsoid body from an anterior and posterior primordium
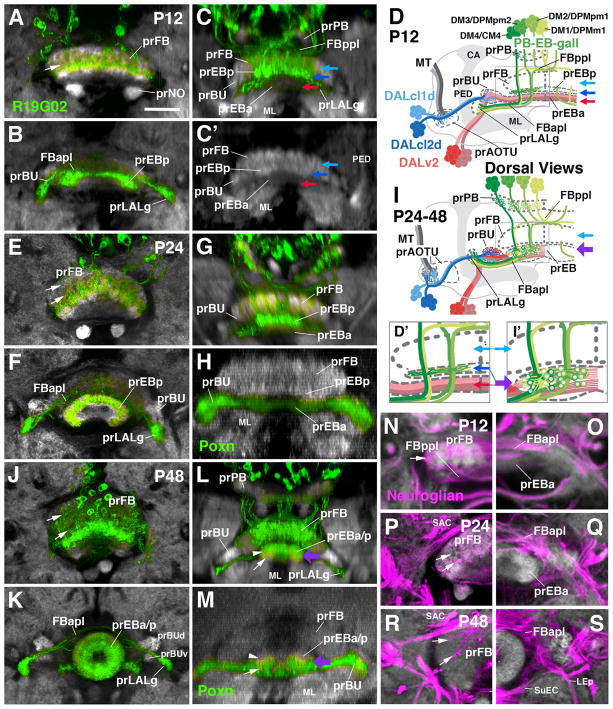
As described above, in the late larva, the axon bundles of the four clusters of R19G02-Gal4-positive PB-EB-gall neurons project from posteriorly into the primordium of the fan-shaped body where they contribute a discrete, ventral layer, the prEBp (see Fig. 2). Around 12h APF, the prEBp separates from the remainder of the prFB (Fig. 5A–D). At this stage one can discern a thick dorso-posterior layer, the prFB (light blue arrow in Fig. 5C, C′, D, D′), and a thinner ventro-anterior layer (prEBp; dark blue arrow in Fig. 5C, C′, D, D′), which will later become incorporated into the ellipsoid body. The prEBp is directly juxtaposed to the anteriorly adjacent prEBa, formed by the distal fibers of DALv2 (red arrow in Fig. 5C, C′, D, D′). Between P12 and P24, the prEBp moves anteriorly and becomes further separated from the prFB (Fig. 5E–I′). At that stage it appears as a curved, DN-cadherin-rich bar that is filled with filopodial endings of R19G02-Gal4-positive PB-EB-gall neurons (Fig. 5F). At P24, the prEBp and prEBa are still largely non-overlapping: R19G02-Gal4 label is restricted to the DN-cadherin-rich prEBp (Fig. 5D, G), and DALv2 markers (e.g., poxn-Gal4) to the DN-cadherin-poor prEBa (Fig. 5H). By P48, as the anterior and posterior primordia of the ellipsoid body merge into one compartment (thick purple arrow in Fig. 5I, I′, L, M), this separation has all but disappeared. R19G02-Gal4-positive fibers have expanded through most of the prEB, with the exception of the anterior domain (arrow in Fig. 5L), confirming the findings of Wolff et al. (2015) who concluded that 19G02-positive PB-EB-gall neurons (“wedge neurons”) are confined to the posterior shell and medial shell of the EB. R-neurons also fill most of the volume of the prEB at P48, leaving only a narrow posterior shell (the future EBop domain; Omoto et al., 2017) empty (arrowhead in Fig. 5M). The subdivision of the EB neuropil into its five distinct domains with different DN-cadherin expression levels (EBa, EBoc, EBic, EBop, EBip; Omoto et al., 2017) becomes apparent only around the time of eclosion.
Figure 5.
Formation of the ellipsoid body from an anterior and posterior primordium. Panels of the left column (A, B, E, F, J, K) show Z-projections of frontal confocal sections of pupal brains in which a subset of columnar neurons of the central complex are labeled by R19G02-Gal4 (green); neuropil is labeled by anti-DN-cadherin (white). Middle column (C, C′, G, H, L, M) presents digitally-tilted horizontal sections of brains labeled with R19G02-Gal4 (C, G, L) or Poxn-Gal4 (H, M). Upper two rows (A–C′) show 12h pupa (P12), middle two rows (E–H) 24h pupa (P24), and bottom two rows 48h pupa (P48). Frontal sections of left column alternatingly show a more posterior plane, containing the primordium of the fan-shaped body (prFB; A, E, J) and an anterior plane with primordium of the ellipsoid body (prEB) and lateral accessory lobe (prLALg; B, F, K). Panels (D, I) are schematic representations of pupal brain hemisphere viewed from dorsally, illustrating architecture of central complex primordia at an early pupal stage (P12, shown in D) and early-mid pupal stage (P24-48, shown in I). In these schematics, neurons of the DALv2 lineage, forming R-neurons of the ellipsoid body, are rendered in red; DALcl1/2d neurons, connecting anterior optic tubercle with bulb, are in blue; columnar neurons of the posterior type II lineages (PB-EB-gall neurons) are in green. (D′, I′) are enlarged views of boxed areas in (D, I). Light blue arrows in (C, C′, D, D′, I, I′) point at prFB; dark blue arrows in (C, C′, D, D′) indicate prEBp, which at this early stage forms part of the prFB; red arrows in (C, C′, D, D′) indicate prEBa; purple arrow in (I, I′, L, M) indicates prEB after the anterior and posterior domains (prEBa, prEBp) have merged. Panels (N–S) are Z-projections of frontal confocal sections of pupal brains (N, O: P12; P, Q: P24; R, S: P48) labeled with anti-Neuroglian (secondary axon tracts; magenta) and anti-DN-cadherin (neuropil; white). Left column (N, P, R) represent posterior plane, containing left half of fan-shaped body primordium (prFB); Right panels (O, Q, S) show anterior planes with right halves of ellipsoid body primordium (prEB). Arrows in (N, P, R) point at longitudinal bundles of PB-EB-LALg axons penetrating the prFB.
For other abbreviations see List of Abbreviations. Bar: 25μm.
3.6. Development of the LALgall
The spreading and elongation of the forward directed PB-EB-gall fibers that terminate in the gall of the LAL can also be followed in pupal brains expressing R19G02-Gal4. At P6 and P12 these fibers penetrate through a central, DN-cadherin-poor layer of the prFB (arrow in Fig. 5A, N). Extending forward they converge in the anterior plexus of the fan-shaped body (FBapl), from which the primordia of the LAL gall (prLALg) protrude as two lateral processes (Fig. 5B, C, D). These primordia are directly adjacent to the anterior lateral ellipsoid body tract formed by the DALv2 (R-neuron) lineage (Fig. 5D). From P24 onward, accompanying the pronounced dorso-ventral growth of the prFB (Fig. 5E), the PB-EB-gall neuronal fiber bundles spread out, forming two tiers of eight tracts each (arrows in Fig. 5E). The primordium of the LALgall adopts its characteristic oval shape and position in the crevice formed by the dorsal surface of the LAL and anterior surface of the bulb (Fig. 5F, I, K).
3.7. Differentiation of neuropil glia during the formation of the AVP and central complex
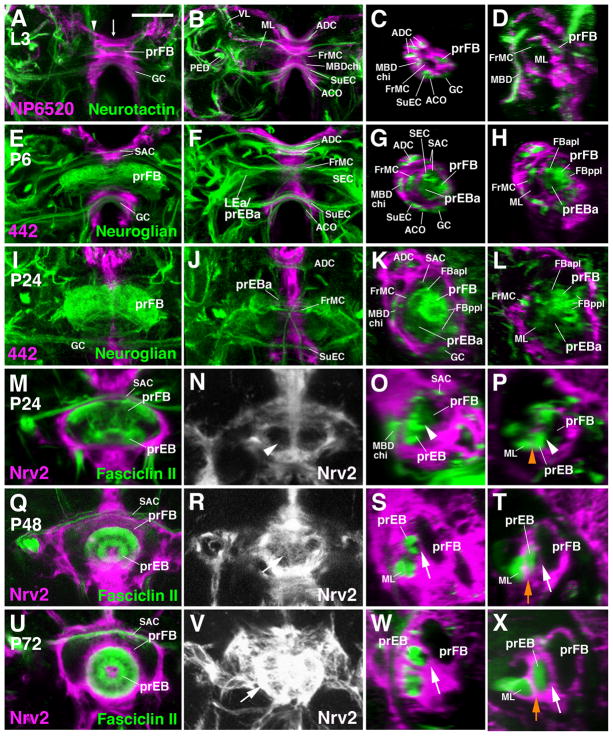
As shown in the previous sections, the primordia of the ellipsoid body and fan-shaped body evolve from a system of closely apposed transverse fiber bundles, formed by several discrete neural lineages. Crossing the midline, these bundles are enveloped by a layer of ensheathing glia (Fig. 6A), and are surrounded on all sides by other commissural tracts, each wrapped into its own coat of ensheathing glia (Fig. 6A–D). The ensheathing glia located at the brain midline represents a small, genetically distinct class of glia that was originally described as the “interhemispheric ring” (Simon et al., 1998). It is labeled by Nrv2-Gal4, a global marker for glia (Younossi-Hartenstein et al., 2003), NP6520-Gal4, a marker for ensheathing glia, (Awasaki et al., 2008), as well as 442-Gal4 (Hitier et al., 2000), whose expression, during pupal stages, is restricted to the interhemispheric ring glia. Using these glial markers in conjunction with BP104 and anti-Fasciclin II we followed the development of glial sheaths as formation of the central complex takes place.
Figure 6.
Development of the glial layer of the central complex. Panels of the first and second column show Z-projections of frontal confocal sections of late larval (A, B) and pupal brains at stages indicated in upper left corner. The third and fourth columns present sagittal sections at the corresponding stages. Panels of the first column (A, E, I, M, Q, U) represent a posterior plane, showing the primordium of the fan-shaped body (prFB); second column panels (B, F, J, N, R, V) are taken at a more anterior plane representing the primordium of the ellipsoid body. Panels of the third column (C, G, K, O, S, W) show sagittal plane right at midline [indicated by arrow in panel (A)], panels of the fourth column (D, H, L, P, T, X) represent a parasagittal plane at approximately 5–10μm lateral of midline [indicated by arrowhead in (A)]. Neuropil glia is labeled by NP6520-Gal4 (A–D), 442-Gal4 (E–L), or Nrv2-Gal4 (M–X; magenta). In all panels except (N, R, V), secondary axon tracts (green) are labeled by anti-Neurotactin (A–D), anti-Neuroglian (E–L) or anti-Fasciclin II (M–X). Processes of neuropil glia forming stable sheaths around individual commissural tracts, or combinations of neighboring tracts, can be recognized in sagittal sections (panels of third column) from larval stages to the late pupa and adult. In the larva and early pupa (C, D, G, H), a common sheath surrounds a system of tracts that includes the primordia of the fan-shaped body (prFB, including posterior primordium of ellipsoid body), anterior ellipsoid body (prEBa), several commissures (supraesophageal commissure, SEC; part of superior arch commissure, SAC) and the tip of the medial lobe of the mushroom body (ML). After P24, a layer of glial processes starts to separate the medial lobe from the ellipsoid body primordium (orange arrow in T, X; orange arrowhead in P, representing 24h pupa, points at position where glial septum will appear at later stages). Concomitantly, glial processes grow in between the primordia of the ellipsoid body and fan-shaped body (white arrows S, T, W, X). Panels (N, R, V) represent the frontal plane that contains the emerging glial sheath (arrowhead in N: no glia present yet; arrows in R, V: glial septum has formed) between ellipsoid body and fan-shaped body primordium; note increasing intensity of the Nrv2 signal, attesting to the growth in thickness of the glial layer.
Other abbreviations: see List of Abbreviations. Bar: 25μm.
During the first 12h of metamorphosis, the primordium of the central complex grows in volume, but remains enclosed in an undivided glial cavity. As described in the previous section, the primordium of the anterior ellipsoid body (prEBa) crosses the midline, and the primordia of the posterior ellipsoid body and fan-shaped body become spatially separated and grow in volume during this time interval, but both remain in direct contact, with no intervening glial layer (Fig. 6E–H). Between 24h and 36h APF, the primordia of the ellipsoid body merge together and adopt their elliptical shape, while still growing in volume. Still, the entire prEB and prFB, and at this stage even the medial lobe of the mushroom body, are enclosed in a shared glial covering (Fig. 6I–P). Around 48h APF, glial processes, formed by secondary ensheathing glia born in the larva (Omoto et al., 2015), start to grow in between the prFB and prEB, as well as between the prEB and the medial lobe (Fig. 6P, T, X; orange arrowhead/arrow). By 72h APF, a thick, largely uninterrupted layer of ensheathing glia surrounds each individual compartment of the central complex and the mushroom body (Fig. 6U–X).
3.8. Projection of primary neurons generated by AVP lineages
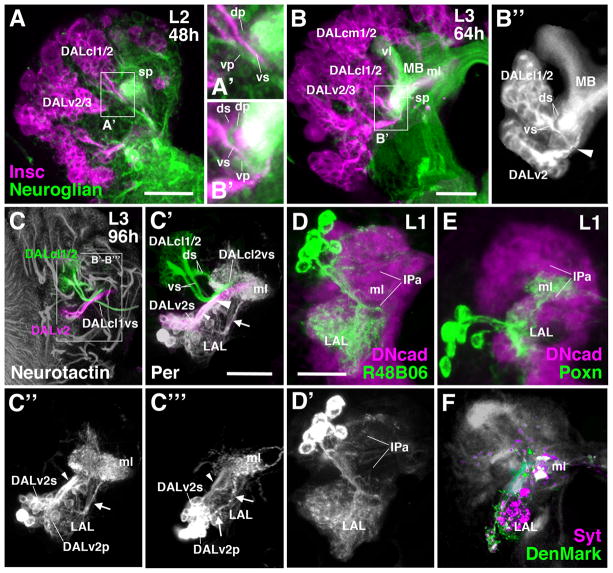
Secondary neurons forming the central complex and AVP are born and extend their axons during the larval period. In general, secondary axon tracts fasciculate with the existing tracts extended (during embryogenesis) by the corresponding primary neurons (Larsen et al., 2009; Hartenstein et al., 2015). In this manner, primary axons of the type II lineages DM1-4 pioneer the primordium of the fan-shaped body (Riebli et al., 2013; Hartenstein et al., 2015). Primary tracts also precede the formation of the AVP. As shown in Fig. 7A/A′ and 7C/C′ for the late second instar larva (L2), primary axons of lineages DALcl1 and DALcl2 (labeled by anti-Neuroglian; green in Fig. 7A/A′, B/B′) approach the mushroom body spur from laterally and split into a dorsal bundle and ventral bundle. Secondary axon tracts of these lineages (labeled by insc-Gal4; magenta in Fig. 7A/A′, B′/C) adopt the same pattern. The dorsal tract is formed by the two dorsal hemilineages, DALcl1d and DALcl2d (Lovick et al., 2013), which represent the TuBu neurons of the anterior visual pathway described in this paper. We speculate that the primary DALcl1/2 neurons contributing to the dorsal tract may similarly represent hemilineages. The same applies for the neurons projecting ventrally. Here, secondary axons of the DALcl1/2v hemilineages form a ventro-medially directed tract that splits into two branches. DALcl1v continues medially and eventually crosses the midline in the SuEC commissure (Lovick et al., 2013; Fig. 7C); DALcl2v turns postero-dorsally, passes underneath the medial lobe of the mushroom body, and approaches the midline at the level of the fan-shaped body primordium (Fig. 7C/C′). Secondary DALcl2v neurons form a major subset of large-field fan-shaped body neurons (Wong et al., 2013). As shown in Fig. 7A, the secondary DALcl1/2v tract is well established 48h after hatching and extends along its primary counterpart. The secondary DALcl1/2d hemilineages differentiate slightly later; they are present 64h after hatching and extend their axons along the primary DALcl1/2d tract (Fig. 7B–B″).
Figure 7.
Elements of the AVP in the early larva. (A–B″) Z-projections of frontal confocal section of larval brain hemisphere labeled with Insc-Gal4>UAS-mcd8GFP (magenta; secondary neurons) and anti-Neuroglian (green; primary neurons) at the L2 stage (48h post hatching; A, A′) and early L3 stage (64h post hatching; B–B″). Note close association of primary and secondary fiber bundles of DALcl1 and DALcl2 (DALcl1/2dp and DALcl1/2ds; DALcl1/2vp and DALcl1/2vs) in (A′) and (B′), respectively. (B″) shows enlarged view of early secondary tracts of DALcl1/2 and DALv2, labeled by Insc-Gal4>UAS-mcd8GFP (white; all other labeled lineages were digitally erased; mushroom body (MB) shown as reference). (C–C‴) Frontal confocal sections of late third instar larval brain hemisphere labeled with anti-Neurotactin (secondary neurons) and Per-Gal4>UAS-mcd8GFP (labels secondary and primary neurons of the DALv2 lineage). In (C), only the Neurotactin labeling is shown; secondary tracts of lineages DALcl1/2 and DALv2 were digitally rendered green and magenta, respectively. (C′–C‴) show Per-Gal4 labeling of both primary neurons (DALv2p) and secondary neurons (DALv2s) of the DALv2 lineage. In (C′), Neurotactin labeling of secondary DALcl1/2 tract and DALv2 tract (digitally rendered green and magenta, respectively) is left; Neurotactin signal of all other secondary neurons was digitally erased. (C″) and (C‴) show Per-Gal4> mcd8GFP in DALv2 in brain of two different L3 specimens. Small arrowhead in (C′–C‴) indicates secondary DALv2 tract (DALv2s); large arrowhead in (C′) and (B″) points at convergence of secondary tracts of DALv2 and DALcl2v. Arrow in (C′–C‴) indicates primary DALv2 tract (DALv2p). (D–E) Z-projections of frontal confocal section of first instar larval brain hemisphere, showing labeling of primary neurons of DALcl1/2d [R48B06-Gal4>UAS-mcd8GFP in (D)] and DALv2 [Poxn-Gal4>UAS-mcd8GFP in (E)]. Antibody against DN-cadherin labels neuropil (magenta). (F) Z-projection of frontal confocal section of late third instar larval brain hemisphere. Constructs labeling postsynaptic sites (DenMark; green) and presynaptic sites (Syntaxin; Syt; magenta) were driven by DALv2 driver Poxn-Gal4. Bars: 20μm (A, B, D–E); 50μm (C–C‴)
In contrast to DALcl1/2 and all other brain lineages analyzed to date, the secondary tract of DALv2, which configures the primordium of the ellipsoid body and bulb in the late larva, does not fasciculate with the primary DALv2 axons (Lovick et al., 2016). Primary DALv2 axons enter the LAL compartment at its anterior-ventral surface. The majority of DALv2 primary neurons appear to arborize within the LAL (see below); a subset of primary DALv2 axons continues dorsally along the LAL surface and approaches the medial lobe of the mushroom body (arrow in Fig. 7C′–C‴). The secondary DALv2 tract extends along the lateral surface of the LAL (small arrowhead in Fig. 7C′, C″). At the position where it enters the neuropil it joins the DALcl2v tract (large arrowhead in Fig. 7C′), passing underneath the medial lobe, and turning medially right posterior to the lobe.
Given that the AVP develops during metamorphosis, where do primary neurons of DALcl1/2d and DALv2 project in the larval brain? The markers R48B06-Gal4 (DALcl1/2d), R82E10-Gal4 (DALcl1d/v), and Poxn-Gal4 (DALv2) are already expressed in the early larva and allowed us to address this question. R48B06-Gal4 labels a cluster of approximately eight neurons which project along the DALcl1/2d tract, crossing the peduncle at its dorsal surface (Fig. 7D, see above). Axons arborize in the LAL, as well as the anterior inferior protocerebrum (IPa) that surrounds the medial lobe of the mushroom body (Fig. 7D/D′). Expression of R82E10-Gal4 (DALcl1d/v) confirms this projection pattern (not shown). Primary DALv2 neurons project to the same compartments (Fig. 7E), filling the lateral half of the LAL, and a dense, cuff-shaped domain surrounding the middle of the medial lobe (ml in Fig. 7E). Expressing DenMark and Synaptotagmin in these neurons shows that pre- and postsynaptic sites of primary DALv2 neurons are present in both the LAL and IPa (Fig. 7F). Postsynaptic sites in the latter are most likely contacted by Kenyon cell axons, which would indicate that primary DALv2 neurons form a pathway interconnecting the mushroom body with the surrounding IPa and LAL (Fig. 7E).
4 Discussion
4.1. Morphogenesis of the central complex neuropil
The central complex, defined as a set of compartments with terminal neurites and synapses spanning the brain midline, is an adult-specific structure of the Drosophila brain that differentiates during metamorphosis. Similarly, the anterior optic tubercle and bulb, which provide visual input to the central complex, develop during metamorphosis. The only contribution of embryonically-born, primary neurons that we currently know about are the “fan-shaped body pioneer neurons” that form a thin bundle of undifferentiated, commissural axons visible already in the early larva (Hartenstein et al., 2015), and that will later differentiate into a subtype of columnar neurons of the central complex (Riebli et al., 2013).
At late larval stages, secondary lineages that will give rise to the bulk of columnar and tangential neurons of the central complex make their appearance. These lineages form tracts that associate themselves with the fan-shaped body pioneers. The tracts, and tufts of filopodia emerging from them, form discrete primordia that are recognizable by a high expression level of DN-cadherin and the absence of markers for mature synapses, like Bruchpilot or Synaptotagmin. The combination of high levels of DN-cadherin and missing expression of synaptic markers is a robust criterion to morphologically recognize immature, non-functional primordia of adult neuropil compartments in the larval brain.
The emergence of the central complex architecture that takes place during the first half of pupal development involves several sequential following steps. First, the bilateral primordia of fan-shaped body and ellipsoid body advance towards the midline, resulting in the formation of a bar-shaped neuropil that straddles the midline. This process is accompanied by a widening of the glial sheath surrounding these primordia. Initially, this sheath forms a tight cuff, enclosing only the commissural tract formed by the fan-shaped body pioneers. During the first 12 hours APF the cuff widens, to accommodate both the fan-shaped body primordium and anteriorly adjacent ellipsoid body primordium (see Fig. 6). The cuff enveloping the developing central complex is formed by the interhemispheric ring glia, a subset of the ensheathing glia surrounding the neuropil (Omoto et al., 2015; see Fig. 6). An important (presumably permissive) role of the interhemispheric ring glia can be gleaned from the results of genetic studies where factors expressed in the interhemispheric ring glia are mutated. Examples of such genetic factors are the receptor tyrosine kinase Linotte/Derailed, whose disruption results in structural defects of the central complex, in particular the appearance of clefts in the midline that divide the fan-shaped body and/or ellipsoid body into a paired neuropil (Simon et al., 1998).
The second major event that shapes the central complex consists in the separation of different layers and columns which, at least in part, represent different channels of input and different functionalities. Columns can be recognized in the prFB and prPB as soon as these primordia make their appearance in the larva (Riebli et al., 2013). By contrast, the hemilineages and sublineages of DALcl1/2 and DALv2, forming the anterior optic tubercle, bulb, and ellipsoid body, appear to overlap widely in the respective primordia of these compartments. A sorting out that begins during the first day of metamorphosis results in the appearance of morphologically distinct subdivisions, including the different subdomains of the tubercle (AOTUm, AOTUim, AOTUil, AOTUla, AOTUli, AOTUlp), the three domains of the bulb (BUs, BUi, BUa), and the five ring domains of the ellipsoid body (EBa, EBoc, EBic, EBop, EBip).
We speculate that repulsive and adhesive membrane components, expressed by DALcl1/2 and DALv2 neurons themselves, as well as by surrounding glial cells, control the growth and spatial sorting that takes place in the EB, BU, and AOTU. In other parts of the developing larval and adult brain, roles of the repulsive Slit/Robo, Semaphorin/Plexin, and Ephrin/Ephrin receptor pathways have been analyzed. Robo receptors were found to be required for the formation of the adult central complex (Nicolas and Preat, 2005). Similarly, Semaphorins/Plexins play a role in the primordium of the adult antennal lobe, where Semaphorin 1a is expressed in a gradient and specifies the location and volume of arborization of olfactory afferents, as well as antennal projection neurons (Komiyama et al., 2007). Semaphorin 1a also acts a receptor for repulsive interactions in the emerging layers of the optic lobe medulla (Pecot et al., 2013). In a similar manner, neurites of sublineages of DALv2 might be sorted out by reacting to repulsive cues acting throughout the primordia of the ellipsoid body and bulb. A recent study (Xie et al., 2017) confirms that Semaphorin/Plexin mediated interactions play an important role in the proper layering of the ellipsoid body.
4.2. Ontogeny and phylogeny of the central complex
The central complex and its related neuropils are highly conserved among insects (Loesel et al., 2002). However, developmental studies addressing these brain compartments are rare (for a recent review, see Koniszewski et al., 2016; Boyan and Reichert, 2011). In hemimetabolan taxa all compartments of the central complex seem to be generated during the embryonic period. The formation of the locust fan-shaped body, which has been studied in detail by Boyan and collaborators (Boyan and Liu, 2014; 2016), shows numerous features which are amazingly similar to the corresponding process in Drosophila, in particular when considering the relationship between lineages and columnar architecture (see below). However, in the locust, FB columnar neurons appear at around 50% of embryonic development, whereas they are born during the larval period in Drosophila. We assume the same applies to the lineages forming the anterior visual pathway, but no detailed information exists on the development of these lineages in the locust.
In most holometabolous taxa, larvae hatch without a fully formed central complex. However, a rudimentary, but differentiated fan-shaped body crossing the midline is typically present. In the coleopteran Tenebrio molitor (mealworm), the fan-shaped body, clearly recognizable by its eight vertical modules already existed in freshly hatched larvae (Wegerhoff and Breidbach, 1992). Interestingly, the columnar units are initially formed by regularly spaced branches of large tangential neurons, rather than the large number of small-field columnar neurons that (in Tenebrio as in other insects) innervate the vertical modules of the fan-shaped body. These columnar neurons, along with the ellipsoid body, make their appearance in the late larva, similar to what has been described in Drosophila (Hartenstein et al., 2015; Lovick and Hartenstein, 2015). We speculate that the early differentiating tangential neurons forming the rudimentary fan-shaped body primordium in Tenebrio may be homologous to the primary neurons whose axons assemble into the simple commissural bundle that constitutes the fly fan-shaped body primordium of the early larva (Riebli et al., 2013). However, whereas these early wide-field neurons differentiate and produce regularly spaced side branches that prefigure the columns of the mature fan-shaped body in Tenebrio, such behavior is not displayed by their Drosophila counterparts, which do not start to differentiate prior to metamorphosis (Riebli et al., 2013).
The ellipsoid body (called “lower unit of the central body” in other insects) generally appears later in development than the fan-shaped body (“upper unit of the central body”). In insect species other than higher diptera, the ellipsoid body forms a bar-shaped neuropil layer across the midline anterior-ventrally to the fan-shaped body (Loesel et al., 2002). In flies, the ellipsoid body resembles the bar-shaped lower division of other insects at early stages (P6-P12), but subsequently detaches fully from the fan-shaped body and bends full circle into an ellipse, as described in the present paper. We speculate that a number of Drosophila mutant phenotypes affecting the central complex represent cases of developmental arrest. For example, the ellipsoid body of flies carrying mutations in ellipsoid body open (ebo; Ilius et al., 2007) show a ventral cleft, suggesting that the primordium of this compartment may have started out normally, but was subsequently prevented from completing the curvature.
Columnar and tangential neuronal elements of the central complex in different taxa are probably formed by homologous lineages. Since the bold paper putting forward this hypothesis (Thomas et al., 1984), it is generally assumed that the pattern of neuroblasts and the lineages they form is highly conserved across all insect groups. During the past three decades, not many lineage comparisons were made; the four lineages forming the columnar neurons of the central complex form a notable exception. In the hemimetabolous locust (Schistocerca gregaria), the central complex contains four prominent lineages (W, X, Y, Z) that generate the columnar neurons of the fan-shaped body and protocerebral bridge (Boyan and Williams, 1997). They bear many of the hallmarks of the four lineages DM1-4 which have the same projections in Drosophila. In both species, the four lineages are produced by type II neuroblasts, which first divide to give rise to a type of cell called “intermediate progenitor” (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Boyan et al., 2010). Intermediate progenitors in turn generate ganglion mother cells, followed by neurons. It is also noteworthy that the axonal scaffold built by the neurons of the four lineages bears an extraordinary resemblance between Schistocerca and Drosophila.
We speculate that the lineages generating the AVP in Drosophila will also have counterparts in the locust and other insects. For Schistocerca, so called TL neurons with morphologies very much like that of DALv2 neurons have been described (Müller et al., 1997). Like Drosophila DALv2 neurons, cell bodies of these neurons in Schistocerca are located in the anterior cortex, and extend a cohesive axon bundle towards the lateral triangle (the neuropil corresponding to the bulb in Drosophila; Homberg et al., 2003; Pfeiffer and Homberg, 2014; Omoto et al., 2017). Similarly, two coherent clusters of neurons, TuLAL1a and TuLAL1b, were described that have cell bodies forming groups lateral of the TL neurons. These neurons interconnect the anterior optic tubercle with the lateral triangle/bulb, and are likely to correspond to the Drosophila TuBu neurons, generated by the lineages DALcl1 and DALcl2.
4.3. Forerunners of the central complex in the Drosophila larva
Lineages of the Drosophila brain are comprised of a set of primary neurons that are born and differentiate in the embryo (“primary neurons”), and a larger set of “secondary neurons” that are born in the larva and differentiate during metamorphosis. In general, primary neurons show characteristics that are fundamentally similar to their secondary siblings. For example, most primary neurons of the four mushroom body lineages, or the antennal lobe projection lineages, have dendritic and axonal arborizations that project in a similar manner as the corresponding secondary neurons (Lee et al., 1999; Das et al., 2013). By contrast, primary neurons forming the characteristic, highly ordered orthogonal lattice of midline-spanning tangential and columnar elements seen in the protocerebral bridge, fan-shaped body, and ellipsoid body do not exist in the larva. This raises the question of where the primary neurons of lineages that form the adult central complex project in the embryo. Furthermore, can one consider this larval neuropil domain innervated by primary neurons of central complex-associated lineages as a precursor of the adult central complex?
We propose that the lateral accessory lobe (LAL), which is an easily recognized part of both larval and adult brain, plays an important role in the context of understanding the emergence of the adult central complex. The LAL has been long recognized as a major output region of the protocerebrum that projects towards motor centers in the subesophageal and thoracic ganglia (Namiki and Kanzaki, 2016). Long projection neurons with arborizations in the LAL can be backfilled when applying tracer molecules to the VNC (Cardona et al., 2009). In the adult brain, neurons of lineage DALd possess the exact same structure (Wong et al., 2013), and we speculate that primary neurons of the DALd lineage have a similar projection (Fig. 8A, B). The adult LAL (specifically: the gall of the LAL) is the only compartment known to receive bundled efferent projections from the columnar neurons of the central complex (Hanesch et al., 1989; Wolff et al., 2015). Thus, when considering the pathway by which the electric patterns generated in the central complex, such as recorded during the control of sleep (Pimentel et al., 2016), aggression (Alekseyenko et al., 2013), body orientation (Strauss, 2002; Seelig and Jayaraman, 2015) and many other activities reach the motor centers, the LAL is likely to represent a key node (Namiki and Kanzaki, 2016).
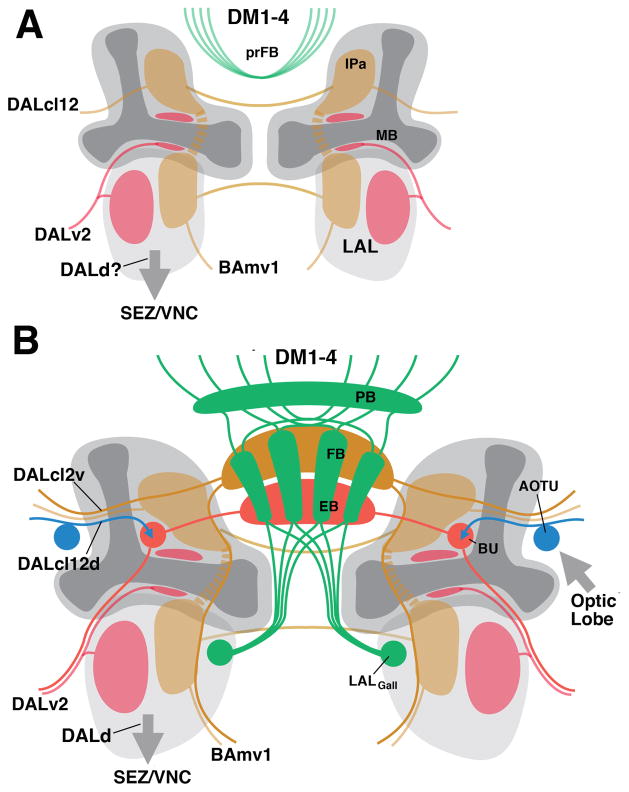
Figure 8.
Schematic representation of projection patterns of lineages building the central complex at the larval stage (A) and adult stage (B).
The LAL also forms dense afferent connections with the central complex. Neurons of lineage BAmv1 have widespread proximal arbors in the LAL, and distal, axonal projections that constitute a type of large-field neuron of the fan-shaped body (Wong et al., 2013; Fig. 8B). The locust lateral triangle, counterpart of the bulb in Drosophila and provider of the visual input to the central complex, is anatomically considered part of the LAL (Homberg et al., 2003). In Drosophila, as shown in this paper, the small neuropil constituting the bulb arises outside, but in direct spatial contact with, the LAL (Fig. 8B)
Interestingly, lineages whose neurons form the AVP of the adult brain also innervate the LAL. Both (secondary) DALcl1 and DALcl2 have dense arborizations in the LAL. DALcl1v neurons form a major source of commissural connections between the LALs of either side (Wong et al., 2013), and the same is true for the primary DALcl1v neurons (Hartenstein et al., 2015). Secondary DALcl2v neurons have sparser projections to the LAL, but interconnect the IPa compartments, located dorsally adjacent to the LAL, and form a class of fan-shaped body tangential neurons (Wong et al., 2013). We speculate that primary DALcl2v and DALcl1v neurons also have properties similar to their secondary siblings, interconnecting the LAL and IPa compartments of either side (Fig. 8A). As to the DALv2 lineage, whose secondary neurons represent the ring neurons of the ellipsoid body: most of its primary neurons also widely arborize in the ipsilateral LAL; a second subset of primary DALv2 neurons forms arborizations in the IPa, in close contact with the mushroom body lobes (Fig. 8A; see also Fig. 7B′). These projections remain intact in the adult brain as well (J.L. and V.H., unpublished observation). Finally, sublineages of some of the type II lineages, notably DM4, which generate the posterior, columnar neurons of the central complex, also densely innervate the LAL, both in the adult brain (Wong et al., 2013) and the larva (J.L. and V.H., unpublished).
We speculate that the LAL and adjacent IPa may fulfill fundamental functions in locomotor control, informing the organism about its location and orientation in space, and adjusting motor output accordingly. A defined set of lineages, including DALcl1/2, DALd, DALv2, DM4 and a few others not mentioned here, scaffold the circuitry of the larval LAL/IPa. For the worm-shaped architecture of the larval body this circuitry is sufficient. To process much more complex tasks, related to the possession of jointed legs and wings for locomotion, and complex sensory arrays like compound eyes and antennae for perceiving the environment, a much larger number of neurons with precisely specified connections would be required. These neurons, constituting the columnar and tangential neurons of the central complex, are added to the existing LAL circuitry by the same lineages that formed the LAL in the first place (Fig. 8B). The central complex could be viewed as an “extension” of the LAL in terms of input and output relationships: the LAL proper still provides much of the input to the central complex via tangential neurons of the fan-shaped body (e.g., lineage BAmv1); in addition, the DALcl1/2d neurons bring in massive visual input via the lateral triangle/bulb, which we would consider a developmentally late add-on to the LAL proper. Output from the central complex reaches another late-forming component of the LAL, the gall.
In conclusion, the central components of the anterior visual pathway (bulb and ellipsoid body), formed by secondary neurons of the lineages DALcl1/2 and DALv2, develop within the territory innervated by primary neurons of these lineages. Bulb and anterior ellipsoid body condense within the IPa, posteriorly adjacent to the medial lobe of the mushroom body. Furthermore, the LALgall, a major output site for the central complex, develops at the dorso-lateral surface of the LAL. One might speculate that primary neurons of the lineages forming the adult central complex build a larval circuit that has a function already resembling that of the adult central complex.
Acknowledgments
This work was supported by NIH grant R01NS096290 to V.H. K.T.N. was supported by the NSF Graduate Research Fellowship Program (No. DGE-0707424). J.J.O. was supported by the Ruth L. Kirschstein National Research Service Award (No.GM007185).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: J.K.L., V.H. Acquisition of data: J.K.L., J.J.O., K.T.N. Analysis and interpretation of data: J.K.L., J.J.O., V.H. Drafting of the manuscript: J.K.L., J.J.O., V.H.
References
- Alekseyenko OV, Chan YB, Li R, Kravitz EA. Single dopaminergic neurons that modulate aggression in Drosophila. Proceedings of the National Academy of Science U S A. 2013;110:6151–6156. doi: 10.1073/pnas.1303446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Drosophila; pp. 214–217. [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. Journal of Neuroscience. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Development. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- Boll W, Noll M. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002;129:5667–5681. doi: 10.1242/dev.00157. [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Developmental Neurobiology. 2008;68:1185–95. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Developmental Cell. 2008;14:535–46. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyan GS, Williams JL. Embryonic development of the pars intercerebralis/central complex of the grasshopper. Development Genes and Evolution. 1997;207:317–329. doi: 10.1007/s004270050119. [DOI] [PubMed] [Google Scholar]
- Boyan G, Williams L, Legl A, Herbert Z. Proliferative cell types in embryonic lineages of the central complex of the grasshopper Schistocerca gregaria. Cell and Tissue Research. 2010;341:259–277. doi: 10.1007/s00441-010-0992-6. [DOI] [PubMed] [Google Scholar]
- Boyan GS, Reichert H. Mechanisms for complexity in the brain: generating the insect central complex. Trends in Neuroscience. 2011;34:247–257. doi: 10.1016/j.tins.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Boyan G, Liu Y. Timelines in the insect brain: fates of identified neural stem cells generating the central complex in the grasshopper Schistocerca gregaria. Development Genes and Evolution. 2014;224:37–51. doi: 10.1007/s00427-013-0462-8. [DOI] [PubMed] [Google Scholar]
- Boyan GS, Liu Y. Development of the Neurochemical Architecture of the Central Complex. Frontiers in Behavioral Neuroscience. 2016;10:167. doi: 10.3389/fnbeh.2016.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Regulation of temporal identities during Drosophila neuroblast lineage development. Current Opinion in Cell Biology. 2005;17:672–675. doi: 10.1016/j.ceb.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Cardona A, Larsen C, Hartenstein V. Neuronal fiber tracts connecting the brain and ventral nerve cord of the early Drosophila larva. Journal of Comparative Neurology. 2009;515:427–440. doi: 10.1002/cne.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Hing H. The L1-CAM, Neuroglian, functions in glial cells for Drosophila antennal lobe development. Developmental Neurobiology. 2008;68:1029–45. doi: 10.1002/dneu.20644. [DOI] [PubMed] [Google Scholar]
- Das A, Gupta T, Davla S, Prieto-Godino LL, Diegelmann S, Reddy OV, Raghavan KV, Reichert H, Lovick J, Hartenstein V. Neuroblast lineage-specific origin of the neurons of the Drosophila larval olfactory system. Developmental Biology. 2013;373:322–337. doi: 10.1016/j.ydbio.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanesch U, Fischbach KF, Heisenberg M. Neuronal architecture of the central complex in Drosophila melanogaster. Cell and Tissue Research. 1989;257:343–366. [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S. The development of the Drosophila larval brain. Advances in Experimental Medicine and Biology. 2008;628:1–31. doi: 10.1007/978-0-387-78261-4_1. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Younossi-Hartenstein A, Lovick JK, Kong A, Omoto JJ, Ngo KT, Viktorin G. Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain. Developmental Biology. 2015;406:14–39. doi: 10.1016/j.ydbio.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitier R, Simon AF, Savarit F, Préat T. No-bridge and linotte act jointly at the interhemispheric junction to build up the adult central brain of Drosophila melanogaster. Mechanisms of Development. 2000;99:93–100. doi: 10.1016/s0925-4773(00)00483-4. [DOI] [PubMed] [Google Scholar]
- Homberg U, Hofer S, Pfeiffer K, Gebhardt S. Organization and neural connections of the anterior optic tubercle in the brain of the locust, Schistocerca gregaria. Journal of Comparative Neurology. 2003;462:415–430. doi: 10.1002/cne.10771. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traguina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Ilius M, Wolf R, Heisenberg M. The central complex of Drosophila melanogaster is involved in flight control: studies on mutants and mosaics of the gene ellipsoid body open. Journal of Neurogenetics. 2007;21:321–338. doi: 10.1080/01677060701693503. [DOI] [PubMed] [Google Scholar]
- Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. Advances in Experimental Medicine and Biology. 2008;628:137–158. doi: 10.1007/978-0-387-78261-4_9. [DOI] [PubMed] [Google Scholar]
- Ito M, Masuda N, Shinomiya K, Endo K, Ito K. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Current Biology. 2013;23:644–655. doi: 10.1016/j.cub.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, Enos A, Hulamm P, Lam SC, Li HH, Laverty TR, Long F, Qu L, Murphy SD, Rokicki K, Safford T, Shaw K, Simpson JH, Sowell A, Tae S, Yu Y, Zugates CT. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. Journal of Comparative Neurology. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nature Reviews Neuroscience. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Sweeney LB, Schuldiner O, Garcia KC, Luo L. Graded expression of semaphorin-1a cell-autonomously directs dendritic targeting of olfactory projection neurons. Cell. 2007;128:399–410. doi: 10.1016/j.cell.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Koniszewski ND, Kollmann M, Bigham M, Farnworth M, He B, Büscher M, Hütteroth W, Binzer M, Schachtner J, Bucher G. The insect central complex as model for heterochronic brain development-background, concepts, and tools. Development Genes and Evolution. 2016;226:209–219. doi: 10.1007/s00427-016-0542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bello B, Reichert H. Lineage-specific cell death in postembryonic brain development of Drosophila. Development. 2008;136:3433–42. doi: 10.1242/dev.037226. [DOI] [PubMed] [Google Scholar]
- Larsen C, Shy D, Spindler S, Fung S, Younossi-Hartenstein A, Hartenstein V. Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Developmental Biology. 2009;335:289–304. doi: 10.1016/j.ydbio.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophial mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Loesel R, Nässel DR, Strausfeld NJ. Common design in a unique midline neuropil in the brains of arthropods. Arthropod Structure and Development. 2002;31:77–91. doi: 10.1016/S1467-8039(02)00017-8. [DOI] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto JJ, Wong DC, Nguyen JD, Hartenstein V. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. Developmental Biology. 2013;384:228–257. doi: 10.1016/j.ydbio.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Hartenstein V. Hydroxyurea-mediated neuroblast ablation establishes birth dates of secondary lineages and addresses neuronal interactions in the developing Drosophila brain. Developmental Biology. 2015;402:32–47. doi: 10.1016/j.ydbio.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Kong A, Omoto JJ, Ngo KT, Younossi-Hartenstein A, Hartenstein V. Patterns of growth and tract formation during the early development of secondary lineages in the Drosophila larval brain. Developmental Neurobiology. 2016;76:434–451. doi: 10.1002/dneu.22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Peña A, Acebes A, Rodríguez JR, Chevalier V, Casas-Tinto S, Triphan T, Strauss R, Ferrús A. Cell types and coincident synapses in the ellipsoid body of Drosophila. European Journal of Neuroscience. 2014;39:1586–601. doi: 10.1111/ejn.12537. [DOI] [PubMed] [Google Scholar]
- Müller M, Homberg U, Kühn A. Neuroarchitecture of the lower division of the central body in the brain of the locust (Schistocerca gregaria) Cell and Tissue Research. 1997;288:159–176. doi: 10.1007/s004410050803. [DOI] [PubMed] [Google Scholar]
- Namiki S, Kanzaki R. Comparative Neuroanatomy of the Lateral Accessory Lobe in the Insect Brain. Frontiers in Physiology. 2016;7:244. doi: 10.3389/fphys.2016.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuser K, Triphan T, Mronz M, Poeck B, Strauss R. Analysis of a spatial orientation memory in Drosophila. Nature. 2008;453:1244–7. doi: 10.1038/nature07003. [DOI] [PubMed] [Google Scholar]
- Nicolaï LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proceedings of the National Academy of Science U S A. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Preat T. Drosophila central brain formation requires Robo proteins. Development Genes and Evolution. 2005;215:530–536. doi: 10.1007/s00427-005-0009-8. [DOI] [PubMed] [Google Scholar]
- Ofstad TA, Zuker CS, Reiser MB. Visual place learning in Drosophila melanogaster. Nature. 2011;474:204–7. doi: 10.1038/nature10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Yogi P, Hartenstein V. Origin and development of neuropil glia of the Drosophila larval and adult brain: Two distinct glial populations derived from separate progenitors. Developmental Biology. 2015;404:2–20. doi: 10.1016/j.ydbio.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Keleş K, Lovick J, Nguyen B, Bolanos C, Frye MA, Hartenstein V. Developmentally- and functionally- distinct parallel neuronal populations relay visual information to the Drosophila central complex in the anterior visual pathway. Current Biology. 2017;27:1098–1110. doi: 10.1016/j.cub.2017.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot MY, Tadros W, Nern A, Bader M, Chen Y, Zipursky SL. Multiple interactions control synaptic layer specificity in the Drosophila visual system. Neuron. 2013;77:299–310. doi: 10.1016/j.neuron.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Kumar A, Jenett A, Reichert H, Hartenstein V. Development-based compartmentalization of the Drosophila central brain. Journal of Comparative Neurology. 2010;518:2996–3023. doi: 10.1002/cne.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K, Homberg U. Organization and functional roles of the central complex in the insect brain. Annual Review of Entomology. 2014;59:165–184. doi: 10.1146/annurev-ento-011613-162031. [DOI] [PubMed] [Google Scholar]
- Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJ, Miesenböck G. Operation of a homeostatic sleep switch. Nature. 2016;536:333–337. doi: 10.1038/nature19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovant M, Léna P. Membrane glycoproteins immunologically related to the human insulin receptor are associated with presumptive neuronal territories and developing neurones in Drosophila melanogaster. Development. 1988;103:145–156. doi: 10.1242/dev.103.1.145. [DOI] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, Taghert PH. Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. Journal of Neurobiology. 1999;41:189–207. [PubMed] [Google Scholar]
- Riebli N, Viktorin G, Reichert H. Early-born neurons in type II neuroblast lineages establish a larval primordium and integrate into adult circuitry during central complex development in Drosophila. Neural Development. 2013;8:6. doi: 10.1186/1749-8104-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig JD, Jayaraman V. Neural dynamics for landmark orientation and angular path integration. Nature. 2015;521:186–191. doi: 10.1038/nature14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AF, Boquet I, Synguélakis M, Préat T. The Drosophila putative kinase linotte (derailed) prevents central brain axons from converging on a newly described interhemispheric ring. Mechanisms of Development. 1998;76:45–55. doi: 10.1016/s0925-4773(98)00104-x. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Hartenstein V. Bazooka mediates secondary axon morphology in Drosophila brain lineages. Neural Development. 2011;6:16. doi: 10.1186/1749-8104-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ. Atlas of an Insect Brain. Springer-Verlag; Berlin: 1976. pp. 1–214. [Google Scholar]
- Strauss R. The central complex and the genetic dissection of locomotor behaviour. Current Opinion in Neurobiology. 2002;12:633–638. doi: 10.1016/s0959-4388(02)00385-9. [DOI] [PubMed] [Google Scholar]
- Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proceedings of the National Academy of Science U S A. 1999;96:10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JB, Bastiani MJ, Bate M, Goodman CS. From grasshopper to Drosophila: a common plan for neuronal development. Nature. 1984;310:203–207. doi: 10.1038/310203a0. [DOI] [PubMed] [Google Scholar]
- Thran J, Poeck B, Strauss R. Serum response factor-mediated gene regulation in a Drosophila visual working memory. Current Biology. 2013;23:1756–63. doi: 10.1016/j.cub.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. Journal of Neuroscience. 2007;27:4541–51. doi: 10.1523/JNEUROSCI.0305-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Wegerhoff R, Breidbach O. Structure and development of the larval central complex in a homometbolous insect, the beetle Tenebrio molitor. Cell and Tissue Research. 1992;268:341–358. [Google Scholar]
- Wolff T, Iyer NA, Rubin GM. Neuroarchitecture and neuroanatomy of the Drosophila central complex: A GAL4-based dissection of protocerebral bridge neurons and circuits. Journal of Comparative Neurology. 2015;523:997–1037. doi: 10.1002/cne.23705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, Hartenstein V. Postembryonic lineages of the Drosophila brain: II. Identification of lineage projection patterns based on MARCM clones. Developmental Biology. 2013;384:258–289. doi: 10.1016/j.ydbio.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Tabuchi M, Brown MP, Mitchell SP, Wu MN, Kolodkin AL. Semaphorin–mediated Lamination Facilitates Inhibitory Synapse Organization and Function in the Drosophila Ellipsoid Body. E-Life. 2017 in press. [Google Scholar]
- Yang JS, Awasaki T, Yu HH, He Y, Ding P, Kao JC, Lee T. Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. Journal of Comparative Neurology. 2013;521:2645–2662. doi: 10.1002/cne.23339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Armstrong JD. Structure of the adult central complex in Drosophila: organization of distinct neuronal subsets. Journal of Comparative Neurology. 2010;518:1500–1524. doi: 10.1002/cne.22284. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra P, Hartenstein V. Early development of the Drosophila brain IV. Larval neuropile compartments defined by glial septa. Journal of Comparative Neurology. 2003;455:435–450. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T. Clonal development and organization of the adult Drosophila central brain. Current Biology. 2013;23:633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]