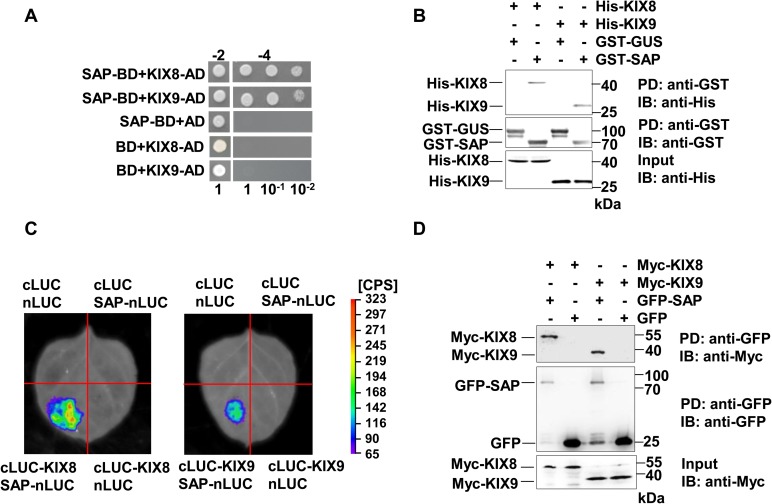
Fig 1. SAP interacts with KIX8 and KIX9.
(A) SAP interacts with full length KIX8 and KIX9 in yeast cells. Transformants were selected on media -2 (SD/-Leu/-Trp), and interactions were tested on media -4 (SD/-Ade/-His/-Leu/-Trp) using a serial dilution of the transformants mixtures (1, 10−1 and 10−2). (B) SAP binds KIX8 and KIX9 in vitro. His-KIX8 and His-KIX9 were incubated with GST-SAP and pulled down by glutathione sepharose. The interactions were detected by immunoblotting with an anti-His antibody. GST-GUS was used as a negative control. (C) The split luciferase complementation assays show that SAP associates with KIX8/9 in N. benthamiana. SAP-nLUC and cLUC-KIX8/9 were co-expressed in N. benthamiana leaves. Luciferase activity was detected 24 hours after infiltration. The pseudocolor bar represents the range of luminescence intensity in each image. (D) SAP interacts with KIX8 and KIX9 in Arabidopsis. GFP-Trap-A beads were incubated with total protein extracts of 35S:GFP;35S:Myc-KIX8, 35S:GFP-SAP;35S:Myc-KIX8, 35S:GFP;35S:Myc-KIX9 and 35S:GFP-SAP;35S:Myc-KIX9 transgenic plants, respectively. The interactions were analyzed by immunoblot with anti-Myc or anti-GFP antibodies.