Figure 37.2.
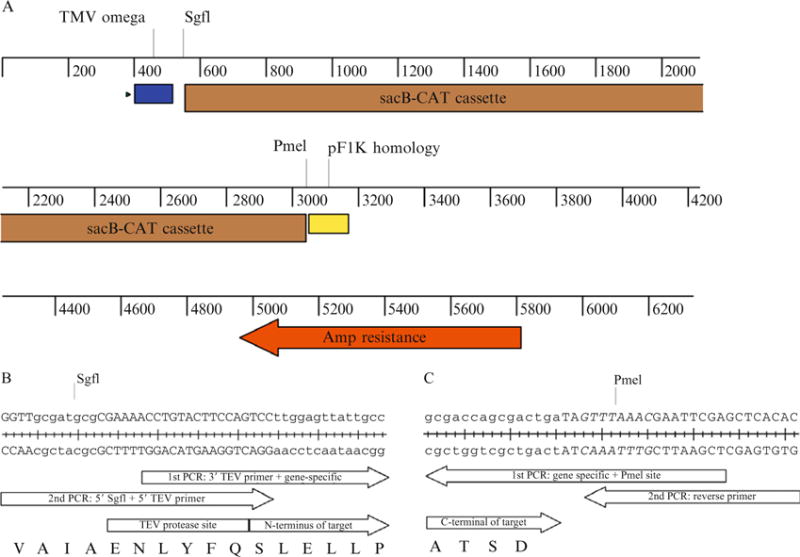
(A) Linear map of pEU-HSCB showing important features of the vector construction. The TMV omega sequence (blue box) enhances translation efficiency. The pF1K homology sequence (yellow box) prevents ligation of two plasmid backbones. (B) Example of the 5′ primer designed for the cloning of Halobacterium salinarium bacteriorhodopsin (GenBank M11720.1). The first-step PCR forward primer is 5′-ACCTGTACTTCCAGTCCttggagttattgcc, with the upper case nucleotides corresponding to the 3′ TEV primer and the lower case nucleotides corresponding to the gene-specific sequence. The second-step universal forward primer is 5′-GGTTgcgatcgcCGAAAACCTGTACTTCCAGTCC, with the SgfI site in lower case. There is an 18 bp overlap between the first-step forward primer and universal forward primer. The TEV protease recognition sequence is ENLYFQ/S, with proteolysis between Q and S. (C) Example of the 3′ primer designed for the cloning of Halobacterium salinarium bacteriorhodopsin (GenBank M11720.1). The first-step PCR reverse complement primer is 5′-GCTCGAATTCGTTTAAACTAtcagtcgctggtcgc, with the upper case, italic nucleotides corresponding to the PmeI site and the lower case nucleotides corresponding to the gene-specific sequence. The second-step universal reverse primer is 5′-GTGTGAGCTCGAATTCGTTTAAAC. There is an 18 bp overlap between the first-step reverse primer and universal reverse primer.
