Extended Data Figure 1. Structures of the DNMT3A–DNMT3L tetramer in complex with the 10/11-mer DNA.
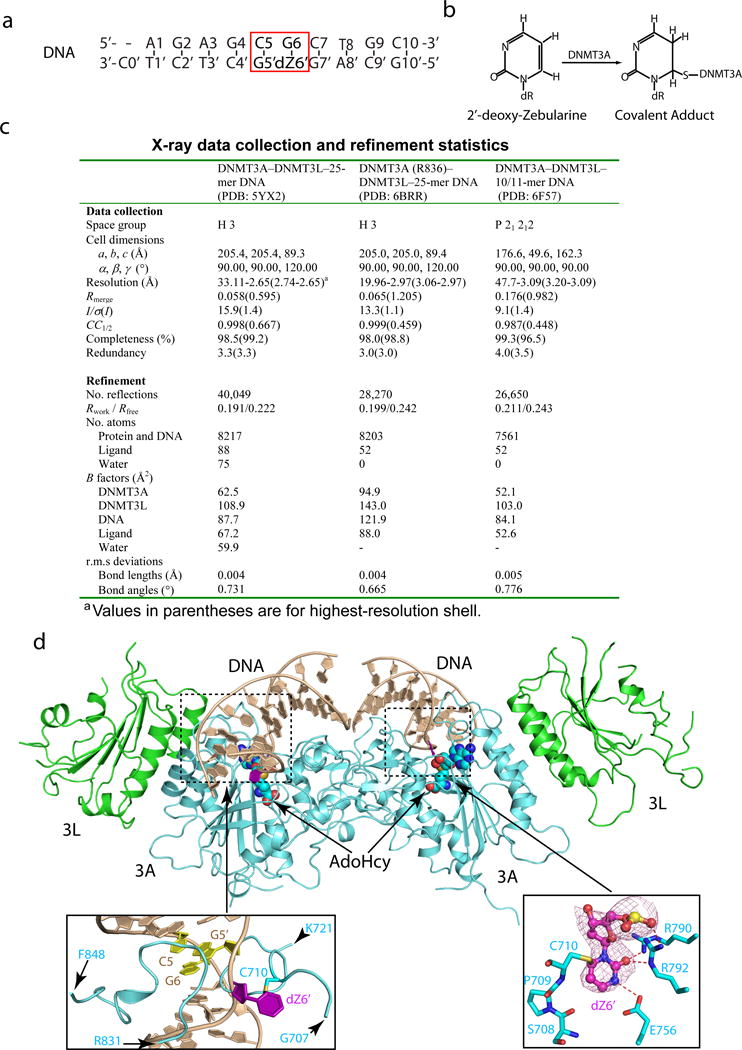
a, The sequence of the 10/11-mer DNA duplex used for structural study. b, Chemical formula of the covalent adduct of DNMT3A and 2′-deoxy-Zebularine. c, Data collection and refinement statistics. Each dataset was collected from a single crystal. d, Ribbon representations of the DNMT3A–DNMT3L tetramer in complex with the 10/11-mer DNA duplex and AdoHcy. DNMT3A, DNMT3L and DNA are colored in light blue, green and wheat, respectively, and AdoHcy shown in sphere representation. The boxed areas show expanded views for the CpG sites (purple and yellow), the DNA-binding TRD and catalytic loops (left box) and the flipped out 2′-deoxy-Zebularine (dZ6′) surrounded by conserved catalytic residues (right box). The Fo–Fc omit map of dZ6′ (pink) is contoured at 3σ level. The hydrogen-bonding interactions are depicted as dashed lines.
