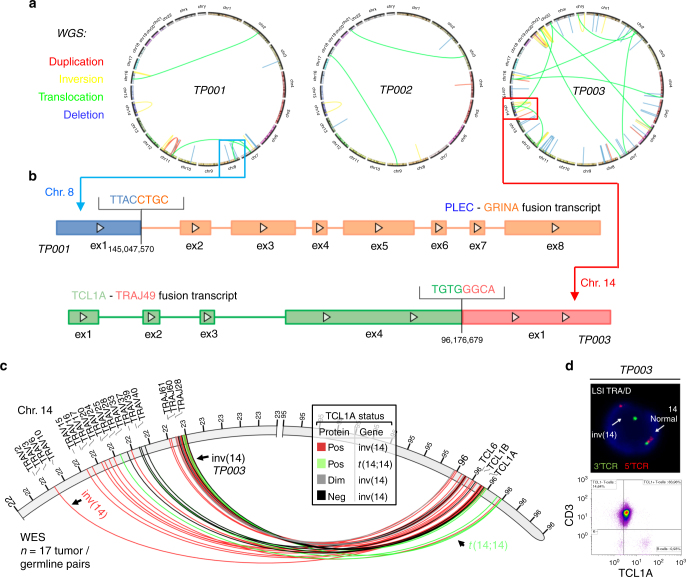
Fig. 3.
Novel structural variations and fusion transcripts. a Whole-genome sequencing (WGS) of 3 T-PLL t/g-pairs to map intra- and inter-chromosomal translocations: 6 lesions affecting 4 distinct chromosomes (TP001), 10 lesions affecting 5 chromosomes (TP002), and 31 lesions affecting 10 chromosomes (TP003); see Supplementary Fig. 8a for whole-exome sequencing (WES) derived data. b Fusion transcripts (n = 96, TopHat-Fusion and oncofuse algorithms) identified by whole-transcriptome sequencing (WTS) of 15 T-PLL compared to healthy donor T-cells (n = 4). Two examples: PLEC-GRINA from aberrations on chr.8 and TCL1A-TRAJ49 (TCRα joining element 49) from an inv(14). c Mapping of breakpoints involved in the inv(14) or t(14;14) derived from WES data on 17 t/g-pairs. Four cases carried each 2 distinct breakpoints. d The FISH-confirmed inv(14) of TP003 (see (b); TCL1A-TRAJ49) was associated with intermediate-level TCL1A protein expression (flow cytometry). Further validation is provided in Supplementary Fig. 8b with proof of a viable transcript and co-expression of neighboring TCL1B