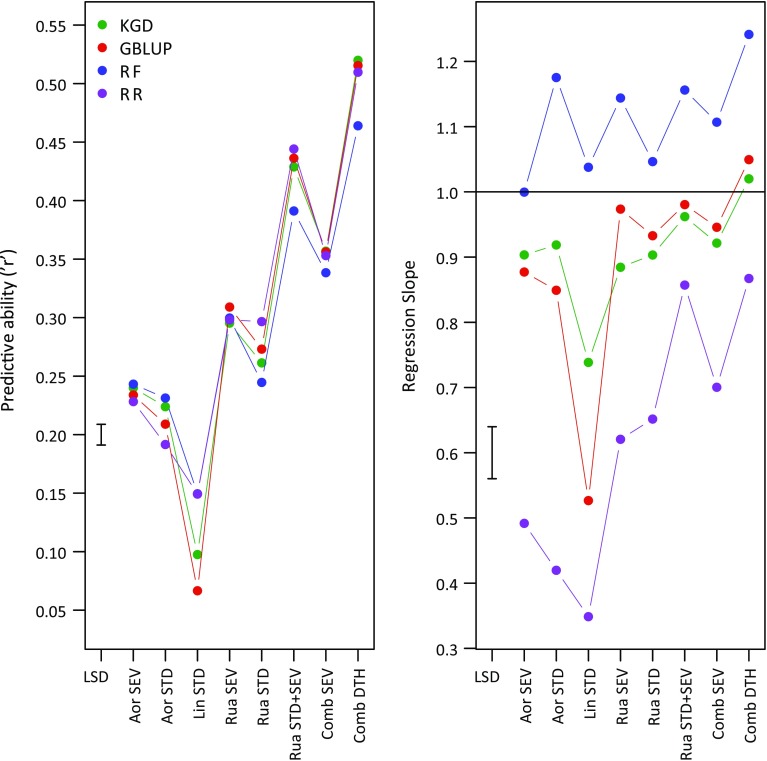
Fig. 3.
Mean (n = 5) tenfold cross-validation predictive ability in a multi-population training set (MP) for seven HA traits and DTH, determined using four statistical models and assessed as a Pearson’s correlation between observed phenotype (BLUP) and GEBV; b slope of the regression of GEBVs on BLUPs. RR ridge regression, RF random forest, KGD GBLUP using KGD genomic relationship matrix. RR models used 249,546 SNPs (largest SNP dataset able to be dealt with computationally by this method), while GBLUP, KGD and RF used 1,023,011 SNPs. Differences between any two statistical methods for each trait > LSD bar are significant at P < 0.05. In b statistical methods with a slope of regression of GEBVs on BLUP-adjusted means ≈ 1 are regarded as providing unbiased estimates of BLUPs. Lines are used on the plots not to infer continuity between the points but to clearly illustrate differences amongst the statistical methods