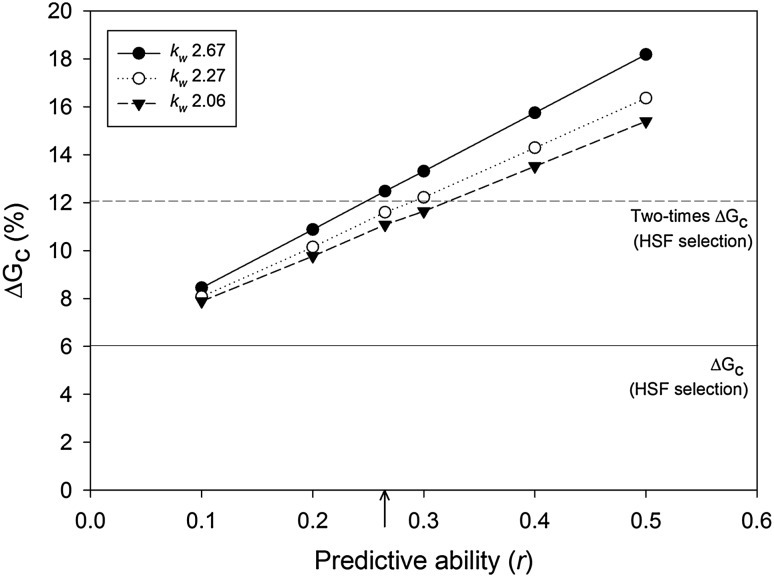
Fig. 5.
Predicted rates of genetic gain (∆G c) for HA based on data from the evaluation of 108 half-sibling (HS) families of perennial ryegrass Pop II in the Rua STD environment. HS family selection (HSF) is compared with among-HS family phenotypic selection and within-family genomic selection (APWFgs-HS). ∆G c is estimated at six levels of genomic-predictive ability (r), including the value estimated by cross-validation for Rua STD HA in Pop II (0.27, indicated by arrow). Three within-HS family selection intensities (k w = 2.06, 2.27 and 2.67, equivalent to selecting the top 5, 3 and 1% of individuals, respectively) are tested at each r. Among-HS family selection intensity is fixed at k f = 1.40 (equivalent to selecting top 20% of HS families) for all HSF and APWFgs-HS scenarios. The solid horizontal line indicates ∆G c for HSF selection (6.02) and the dotted line shows two times that rate (12.04)