Summary
Rust fungi are devastating plant pathogens and cause a large economic impact on wheat production worldwide. To overcome this rapid loss of resistance in varieties, we generated stable transgenic wheat plants expressing short interfering RNAs (siRNAs) targeting potentially vital genes of Puccinia striiformis f. sp. tritici (Pst). Protein kinase A (PKA) has been proved to play important roles in regulating the virulence of phytopathogenic fungi. PsCPK1, a PKA catalytic subunit gene from Pst, is highly induced at the early infection stage of Pst. The instantaneous silencing of PsCPK1 by barley stripe mosaic virus (BSMV)‐mediated host‐induced gene silencing (HIGS) results in a significant reduction in the length of infection hyphae and disease phenotype. These results indicate that PsCPK1 is an important pathogenicity factor by regulating Pst growth and development. Two transgenic lines expressing the RNA interference (RNAi) construct in a normally susceptible wheat cultivar displayed high levels of stable and consistent resistance to Pst throughout the T3 to T4 generations. The presence of the interfering RNAs in transgenic wheat plants was confirmed by northern blotting, and these RNAs were found to efficiently down‐regulate PsCPK1 expression in wheat. This study addresses important aspects for the development of fungal‐derived resistance through the expression of silencing constructs in host plants as a powerful strategy to control cereal rust diseases.
Keywords: host‐induced gene silencing, PsCPK1, wheat, Puccinia striiformis f. sp. tritici, virulence, RNAi
Introduction
Wheat stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the most serious diseases of wheat (Triticum aestivum) worldwide (Chen et al., 2013). This disease can result in more than 90% yield losses in a field (http://striperust.wsu.edu). The most economical, effective and environmentally friendly way of controlling this disease is to breed and use wheat varieties. However, most race‐specific host resistance genes have transient protection, probably due to the rapid evolution of new virulent rust fungal isolates (Fisher et al., 2012). Therefore, new feasible methods must be uncovered to protect wheat crops from rust fungi.
RNA interference (RNAi) is firstly discovered in Caenorhabditis elegans (Fire et al., 1998), and it can trigger potent and specific interference of pathogenic processes through exogenous double‐stranded RNA (dsRNA). For several insects and nematodes, silencing has been confirmed by merely feeding the dsRNA of the target genes (Hannon, 2002; Huvenne and Smagghe, 2010). With the identification of small RNAs (siRNAs), the mechanism of gene silencing of essential cellular functions was established (Castel and Martienssen, 2013; Vaucheret and Fagard, 2001). RNAi has been proved to be a powerful tool to reveal functions of target genes in organisms (Hellens et al., 2005) and offers an effective strategy to enhance resistance in crop plants. The accumulation of RNAi molecules in barley targeting fugal transcripts affects the development of Blumeria graminis f. sp. hordei in barley (Nowara et al., 2010). Transient silencing genes encoding MAP kinases, calcineurin B (PtCNB) and cyclophilin (PtCYC1), lead to increased resistance in wheat against rust diseases (Panwar et al., 2013a). Expressing inverted repeat fragments of cellulose synthase (CES1) genes of Bremia lactucae in transgenic plants resulted in attenuated pathogenicity and growth of B. lactucae (Govindarajulu et al., 2015). Three RNAi constructs derived from Chs3b were identified as the most effective RNAi constructs for enhancing resistance to Fusarium pathogens in planta (Cheng et al., 2015).
As a biotrophic parasite, Pst infects the host mainly from urediospores, which germinate within 3 h at a low temperature after deposition on the leaf surface (Hassebrauk and Schroeder, 1964). Germ tubes grow perpendicular to the long axis of epidermal cells of the leaf until they encounter a stoma. At 6–8 h postinoculation (h p.i.), an appressorium forms above the stoma and subsequently a substomatal vesicle forms within the stomatal cavity. At 12–18 h p.i., the primary infection hypha and haustorial mother cells emerge. Haustorial mother cells (HMCs), which have a thick, multilayered wall, invaginate the host cell plasma membrane with a slender neck to form the haustorium (Kang et al., 2003). Haustoria withdraw nutrients from host cells through the extrahaustorial matrix (Voegele and Mendgen, 2003). The primary infection hyphae branch and produce a number of HMCs and haustoria from 24 to 144 h p.i., developing into the fungal mycelium within the host tissue. From 6 to 8 days after infection, symptoms of chlorosis will be observed, whereas sporulation commences after approximately 12–14 days under favourable conditions.
The cyclic adenosine monophosphate protein kinase A (cAMP‐PKA) signalling pathway is well conserved across eukaryotes and has been proved to participate in virulence, morphogenesis and development in diverse fungi (Bahn and Sundstrom, 2001; D'Souza and Heitman, 2001; Fuller and Rhodes, 2012). In Saccharomyces cerevisiae, cAMP‐dependent protein kinase has a vital role in controlling proliferation, stress resistance, metabolism and the availability of nutrients (Thevelein and De Winde, 1999; Toda et al., 1987). The catalytic subunits of PKA including Tpk1‐3 have different functions. Tpk2 has a unique role in the activation of pseudohyphal growth. In contrast, Tpk1 and Tpk3 repress filamentation (Pan and Heitman, 1999; Robertson and Fink, 1998). Further studies revealed that Tpk2 has a negative role in regulating genes for iron uptake, whereas it has a positive role in regulating genes for water homeostasis and trehalose degradation (D'Souza et al., 2001). In Magnaporthe oryzae, CPKA was shown to inhibit appressorium formation and the responsiveness of germinating conidia to exogenous cAMP (Mitchell and Dean, 1995). The maintenance of pathogenicity in cpkA mutants on wounded plants implies an additional role of CPKA that may be essential for appressorial penetration (Xu et al., 1997). In Fusarium graminearum, CPK1 is responsible for hyphal growth, differentiation and pathogenesis (Hu et al., 2014). In Ustilago maydis, two genes adr1 and uka1 are found to encode catalytic subunits of PKA. The adr1 is the major PKA catalytic subunit gene and required for pathogenicity, whereas uka1 has almost no influence on pathogenicity (Dürrenberger et al., 1998). The availability of rust fungus genomic resources (Duplessis et al., 2011; Xu et al., 2011; Zheng et al., 2013) accelerates the prediction of a great number of genes and the research on functional genomics of rust fungi. However, due to the lack of functional genomics tools for rust fungi, less is known about biological functions of these genes.
In this study, we characterized a gene encoding the catalytic subunit of PKA, designated PsCPK1 in Pst. The results showed that knockdown of PsCPK1 leads to decreased virulence of Pst. The hairpin silencing constructs of PsCPK1 expressed in wheat plants are sufficient to suppress disease development of Pst, indicating durable resistance at the genetic level against Pst infection.
Results
Pst contains two PKA catalytic subunit genes
A BLAST search using adr1 and uka1 of U. maydis (Dürrenberger et al., 1998) as queries revealed that the genome of Pst contains two PKA catalytic subunit genes, PSTG06781 and PSTG11839, that were named PsCPK1 and PsCPK2, respectively (Zheng et al., 2013). In this study, sequence analysis indicated that PsCPK1 has an open reading frame (ORF) of 1443 bp, encoding a putative protein composed of 480 amino acids with a molecular weight of 55.69 kD and an isoelectric point (pI) of 6.72. A multisequence alignment with seven CPK proteins of different organisms in NCBI database revealed that PsCPK1 is 80% and 57.3% identical to CPK1 from Puccinia graminis f. sp. tritici (Pgt) and Puccinia triticina (Pt), respectively, and contain almost all the conserved domains of the PKA catalytic subunit. Compared to the PsCPK1 protein, PsCPK2 shares 30.57% similarity and 40.54% identity at the nucleotide sequence level. Phylogenetic analysis indicated that the homologs of CPK in Pst separate into two distinct groups, class I and class II (Figure 1). PsCPK1 in class I is orthologous to adr1 of U. maydis, cpkA of M. oryzae and yeast TPK2, whereas PsCPK2 in class II is orthologous to cpk2 of M. oryzae and uka1 of U. maydis. These results indicate that CPK is highly conserved in other filamentous fungi.
Figure 1.
Phylogenetic analysis of the catalytic subunits of fungal cAMP‐dependent protein kinases. The amino acid sequences encoded by the catalytic subunits of the cAMP‐dependent protein kinases from Puccinia striiformis f. sp. tritici (PSTG), P. triticina (PTTG), P. graminis f. sp. tritici (PGTG), Ustilago maydis (UM), Cryptococcus neoformans (CNAG), Fusarium graminearum (FGSG), Magnaporthe oryzae (MGG), Saccharomyces cerevisiae (SCRG), Neurospora crassa (NCU) and Aspergillus nidulans (AN) were retrieved from the NCBI. Phylogenetic analysis was carried out with the MEGA6 software by the neighbour‐joining method.
PsCPK1 is highly expressed at the early infection stage of Pst
To investigate whether PsCPK1 is involved in Pst infection, quantitative RT‐PCR (qRT‐PCR) was used to test PsCPK1 transcript levels in different Pst infection stages. The transcript level of PsCPK1 was gradually induced as early as 6 h p.i. and at 18 h p.i. attained the maximum level of 11.7‐fold compared with that in the control. Then, the transcript level returned to the original level at 24–48 h p.i. (Figure 2). Its expression down‐regulated between 72 and 264 h p.i. and was barely detected at the sporulation stage (216–264 h p.i.). Our results indicate that the transcription of PsCPK1 is induced during the infection stage.
Figure 2.
Transcript profiles of PsCPK1 at different Pst infection stages. Wheat leaves inoculated with virulent Pst isolate CYR32 sampled at 0, 6, 12, 18, 24, 36, 48, 72, 120, 144, 216 and 264 h postinoculation (h p.i.). Relative expression of PsCPK1 was calculated by the comparative threshold (2−ΔΔ CT) method. Mean and standard deviation were calculated with data from three independent biological replicates. Differences were assessed using Student's t‐tests. Asterisks indicate P < 0.05, and double asterisks indicate P < 0.01.
PsCPK1 partially complements the M. oryzae cpkA mutant
The PsCPK1 protein shares 80% similarity and 66% identity with CPK1 of M. oryzae. To perform complementation analysis, we transformed the PsCPK1 gene into the cpkA mutant DF51. The resultant transformants showed identical phenotypes and only transformant CM‐41 is used for subsequent analysis. On oatmeal agar medium plates, aerial mycelium of transformant CM‐41 was similar to that of Guy11 (the wild‐type strain), and almost no aerial mycelium was observed in the cpkA mutant DF51 (Figure 3a). These results indicate that the PsCPK1 gene partially complements the defects of the M. oryzae cpkA mutant (DF51) in vegetative growth.
Figure 3.
Complementation of the cpkA mutant with the PsCPK1 fusion construct. (a) Colony morphology of Magnaporthe oryzae strains. Colonies of the wild‐type (Guy11), cpkA deletion mutant (DF51) and complemented strain (CM‐41) grown on PDA plates for 5 dpi. (b) Appressorium formation assay. Germ tubes from the wild‐type strain (Guy11) developed appressoria by 24 h p.i., but no appressorium formation was observed in the cpkA mutant DF51. Under the same conditions, a transformant of expressing the PsCPK1 fusion construct (CM‐41) formed appressoria. Bar, 25 mm. (c) Barley infection assay. Left to right, barley leaves were sprayed with sterile water and conidia of Guy11, DF51 or CM‐41. Typical leaves were photographed at 6 dpi postinoculation.
Further assays on appressorium formation and plant infection with the transformant CM‐41 were performed. The results showed that over 90% of the germ tubes formed appressoria by 24 h in Guy11. However, under the same conditions, approximately 50% of the germ tubes formed appressoria in CM‐41 and no appressoria was observed for the cpkA mutant DF51 (Figure 3b). To test the pathogenicity of the transformant CM‐41, eight‐day‐old barley seedlings of cultivar NB6 were sprayed with conidia of CM‐41. At 6 dpi, leaves inoculated with CM‐41 or Guy11 developed typical blast lesions, whereas fewer and smaller lesions were observed on leaves inoculated with CM‐41 (Figure 3c). No lesions were found on leaves sprayed with water or conidia of DF51 (Figure 3c). Our data show that PsCPK1 can partially complement the DF51 in appressorium formation and plant infection.
Transient silencing of PsCPK1 significantly reduces pathogenicity of Pst
BSMV‐mediated HIGS was adopted to silence PsCPK1 in Pst. Two specific fragments of PsCPK1 were designed for silencing this gene (Figure S1). Ten days after infection with BSMV, wheat seedlings inoculated with sterile FES buffer developed normal leaves (Figure 4a). Under the same conditions, photobleaching was observed in BSMV:TaPDS‐as infected plants (Figure 4a), indicating that TaPDS was successfully and specifically silenced. All wheat seedlings inoculated with BSMV:γ (control), BSMV:PsCPK1‐1as and BSMV:PsCPK1‐2as showed mild chlorotic mosaic symptoms on the third leaf (Figure 4a), suggesting that the BSMV‐HIGS system functioned well. In comparison with the control plants, wheat leaves infected with BSMV:PsCPK1‐1as and BSMV:PsCPK1‐2as displayed a susceptible phenotype with similar viral infection symptoms (Figure 4b). However, the number of uredia in BSMV:PsCPK1‐1as‐ and BSMV:PsCPK1‐2as‐infected leaves was significantly lower than that in the control (Figure 4b). Compared with control plants, the ratio of pustules on leaves infected with BSMV:PsCPK1‐1as and BSMV:PsCPK1‐2as was reduced by 49% and 54%, respectively (Figure 4c). To test silencing efficiency of BSMV‐HIGS, qRT‐PCR was used to assay the relative transcript level of PsCPK1 at 24, 48 and 120 h p.i. with the virulent CYR32 isolate. Compared with control plants, the transcript level in the BSMV:PsCPK1‐1as‐infected leaves was reduced by 74%, 85% and 58%, respectively (Figure 4d). Similarly, in BSMV:PsCPK1‐2as‐infected leaves, the transcript level of PsCPK1 was reduced by 75%, 81% and 62%, respectively (Figure 4d).
Figure 4.
Functional assessment of PsCPK1 in Pst pathogenicity determined by BSMV‐mediated HIGS. (a) Mild chlorotic mosaic symptoms were observed on the fourth leaves of wheat seedlings inoculated with BSMV: γ (control), BSMV:PsCPK1‐1as and BSMV:PsCPK1‐2as. No change in phenotype was observed in wheat leaves mock‐inoculated with FES buffer (MOCK). Photobleaching was evident on wheat leaves infected with BSMV:TaPDS‐as. (b) Phenotypes of the fourth leaves of BSMV:γ (control)‐, BSMV:PsCPK1‐1as‐ and BSMV:PsCPK1‐2as‐inoculated wheat plants 14 dpi with Pst isolate CYR32. (c) Quantification of the uredial density in the BSMV:γ‐, BSMV:PsCPK1‐1as‐ and BSMV:PsCPK1‐2as‐inoculated wheat plants 14 dpi with Pst isolate CYR32. (d) Relative transcript levels of PsCPK1 in the BSMV:γ‐, BSMV:PsCPK1‐1as‐ and BSMV:PsCPK1‐2as‐inoculated wheat plants 24, 48 and 120 h p.i. with Pst isolate CYR32. Values are expressed relative to the endogenous Pst reference gene EF1, with the empty vector (BSMV:γ) set at 1. Values represent the means ± standard error of three independent assays. Differences were assessed using Student's t‐tests, and asterisks indicate P < 0.05.
The detailed histological changes in HIGS‐silenced plants inoculated with Pst CYR32 were microscopically examined (Figure S2a–f). In BSMV:PsCPK1‐1as‐ and BSMV:PsCPK1‐2as‐inoculated leaves, the hyphal lengths exhibited significant reduction relative to that in the control plants at 48 and 120 h p.i. (Figure S2g). The infection areas in the silenced plants also decreased at 120 h p.i. (Figure S2h). However, the numbers of haustorial mother cells showed no significant difference at 48 h p.i. (Figure S2i).
Molecular analysis of transgenic wheat plants
To efficiently generate specific siRNAs directed against the PsCPK1 gene in transgenic wheat plants, the coding region of PsCPK1 (1443 bp) was cloned into the plasmid pMCG161 and the cloned fragments as inverted repeats under control of the maize ubiquitin promoter (Ubi1) to generate the dsRNA (Figure 5a). The resultant construct was then bombarded into wheat cultivar (cv.) Xinong1376 (XN1376). In the glasshouse experiments, ten transgenic lines (L1, L2, L5, L9, L10, L11, L12, L14, L17 and L18) (Table S1) were tested and the disease severity scale was evaluated by the standard described in Table S2 (Line and Qayoum, 1992). Of these ten lines, the transgenic L12 and L18 lines were selected for further study (Figure S3). Integration of the transgene in the two lines L12 and L18 carrying the resultant constructs was confirmed by genomic PCR with primers Bar‐F/R, UBI1‐F/R and TG‐CPK1‐F/R (Table S3; Figure 5a). To further verify that these two lines were independently derived and transgenic, Southern blotting was performed with genomic DNA from the two lines digested with BamHI and XhoI. The results showed that each line contained one copy and exhibited a different banding pattern (Figure 5b). To confirm whether HIGS efficiently works to protect transgenic wheat plants from Pst infection, northern blotting was performed to detect PsCPK1 dsRNA expression in L12 and L18. The results show that L12 and L18 produce the small interfering RNAs (siRNAs, ~21 nt) (Figure 5c; Figure S4).
Figure 5.
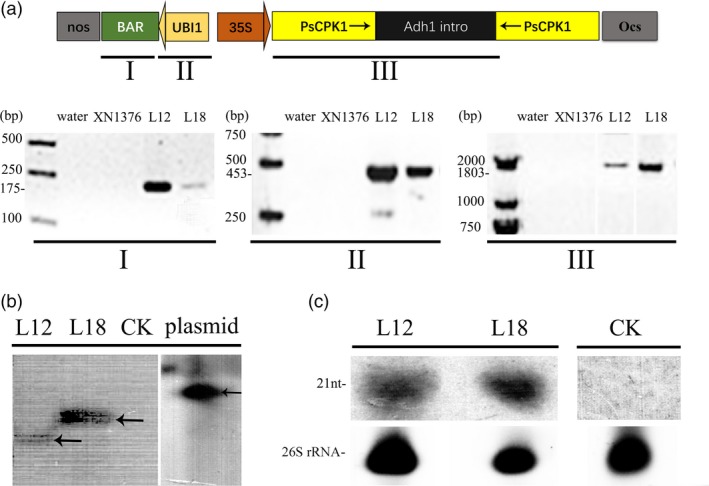
Molecular analysis of the Pst‐resistant transgenic wheat plants. (a) Diagram showing the RNAi cassette in the construct pMCG161‐RNAi for wheat transformation. T4 plants were analysed by genomic PCR for the presence of the selectable marker gene bar (I), UBI1 promoter (II) and fragment (PsCPK1‐intro) of the RNAi cassette (III). The III image was compiled from different pictures. (b) Southern blotting analysis of the transgenic plants. Genomic DNA isolated from control plants (CK), T4 transgenic plants (L12 and L18) and plasmid pMCG161‐PsCPK1‐RNAi. DNAs were digested with Bam HI and XhoI and hybridized to the hairpin fragment probe. The image was compiled from the same picture which we removed the lanes indicating other T4 transgenic lines. (c) The expression of the small RNA in T4 generation lines was analysed by RNA gel blotting. 26S rRNA, loading/blotting control. RNA blots were hybridized with the corresponding target gene‐specific probes. siRNA levels in plants silenced using the pMCG161‐PsCPK1‐RNAi DNA vector expressing target fungal fragments with a mixture of sense plus antisense forms (L12 and L18). No signal was detected in the transgenic lines carrying empty vector (CK). Total RNA was extracted from three different plants. Lower panels, ethidium bromide‐stained rRNA as gel loading controls. The arrowhead indicates an oligonucleotide marker of 21 nucleotides.
Expression of PsCPK1 small RNA in wheat confers durable resistance to Pst
Pst resistance was evaluated in the glasshouse. Average reduction in disease severity in L12 and L18 was 59% and 46%, respectively (Figure S3). Phenotypically, the second leaves of 14‐day‐old L12 and L18 plants were challenged with the virulent CYR32 isolate (Figure 6a). All of the infected leaves could be separated into three classes (Table S2), and the ratio of diseased plants in the 1–3 grade was 69% and 77% in L12 and L18 (Figure 6b). To determine the transcript levels of PsCPK1 at different infection stages of Pst on XN1376, we performed qRT‐PCR assay (Figure S5). According to the results, we isolated total RNA from silenced leaves after 48, 72, 120 and 168 h p.i. with Pst urediospores to test silencing efficiency of PsCPK1. Relative expression analysis of these samples compared with transgenic controls revealed specific reductions in transcript levels of the CPK1 gene in Pst. The transcript levels of the PsCPK1 showed an approximate 42% to 64% down‐regulation in L12 and 36% to 50% in L18 compared with controls (Figure 6c), suggesting that PsCPK1 suppression had an approximate relevance to increased resistance against Pst in L12 and L18. Additionally, a series of experiments provided evidence of the effectiveness and stability of the transgenes. To determine the ratio of sporulating to nonsporulating uredia, a quantitative assay was made within a defined surface area of the leaves at 16 days after Pst infection, which is consistent with the rust disease phenotype. The ratio of sporulating to nonsporulating uredia was significantly reduced by about 80% in transgenic lines. Fungal biomass in the infected leaves was also measured. Total genomic DNA was isolated from wheat leaves infected with Pst, and the relative levels of PsEF1 and TaEF1 were quantified by Q‐PCR (Figure S6). Compared with controls, fungal biomass was significantly reduced by about 76% in L12 and 72% in L18 at 7 dpi, respectively (Figure 6e). Together, these results indicate that in transgenic wheat plants PsCPK1 expression is efficiently down‐regulated and Pst growth is impaired and delayed.
Figure 6.
Expression of PsCPK1 small RNA in wheat confers durable resistance to Pst. (a) Phenotypes of the second leaves of the third‐ and the fourth‐generation wheat plants at 14 dpi with Pst isolate CYR32. (b) The ratio of plants in disease grades. Phenotypes were scored to indicate the frequency of symptoms, as follows: I: 1–3; II: 4–6; III: 7–9. (c) Relative transcript levels of PsCPK1 in the second leaves of the fourth‐generation wheat plants at 48, 72, 120 and 168 h p.i. with Pst isolate CYR32. Values are expressed relative to the endogenous Pst reference gene PsEF1, with the empty vector (CK) set at 1. Values represent the means ± standard error of three independent samples. (d) Quantification of the uredial density in the CK‐, L12‐ and L18‐inoculated wheat plants at 16 dpi. (e) Q‐PCR measurement of fungal biomass. Ratio of fungal to wheat nuclear genomes using fungal PsEF1 and wheat TaEF1 genes, respectively, in plants treated with variants targeting fungal genes compared with controls. Genomic DNA extracted from the second leaf from three different plants at 7 dpi. Values represent the means ± standard error of three independent samples. Differences were assessed using Student's t‐tests. Asterisks indicate P < 0.05, and double asterisks indicate P < 0.01.
Histological and molecular changes in Pst growth in transgenic plants
To further test the possible function of the PsCPK1 in Pst–wheat interaction, we made microscopic assessment of Pst development in foliar tissue. The infection sites were chosen randomly from the second infected leaves of L12 and L18 plants and transgenic control plants at 48 and 120 h p.i. and analysed by fluorescence microscopy. At 48 h p.i., fungal penetration and expansion abilities were not different, and the mycelial morphology appeared normal compared with that in the transgenic control lines (Figure 7a–c). Mycelial morphology appeared normal (Figure 7d–f), whereas at 168 h p.i. both the length of infection hyphae (IH) (Figure 7g) and area of infection unit were significantly reduced (Figure 7h). There were no significant differences in the number of haustorial mother cells at 48 and 168 h p.i. (data not shown). To further understand the relationship between transgenic wheat and controls, we assayed the transcript profiles of a few selected genes after infection with Pst. The transcript levels of PsRAS2, upstream of the cAMP‐PKA pathway in Pst, were down‐regulated in transgenic lines compared with the control (Figure 8), but PsRAS1 was only down‐regulated at 48 h p.i. We also subsequently monitored the relative expression of several other subunit genes of the PKA complex (PsCPK2 and PsRPK) in transgenic plants (Figure 8). Interestingly, both were down‐regulated at 168 h p.i., but showed no obvious changes at 48 h p.i. (Figure 8). PsPrf1, a transcription factor functioning after phosphorylation by PsCPK1, decreased significantly in the L12 and L18 lines at 48 and 168 h p.i. Therefore, our data indicate that PsCPK1 most likely plays an important role in the virulence of Pst by participating in fungal development and growth, and silencing of the PsCPK1 results in virulence penalty of Pst in transgenic plants.
Figure 7.
Histological changes in Pst growth in transgenic plants. (a–f) A microscopic examination revealed no obvious differences in the number of hyphal branches between the control plants (CK) and the transgenic plants (L12 and L18) at 48 h p.i. (a–c) and 168 h p.i. (d–f), respectively. (g) The hyphal lengths in the transgene lines (L12 and L18) were shorter than those observed in the control plants (CK) at 168 h p.i. (h) The colony sizes in the two transgenic plants (L12 and L18) were reduced compared with the sizes observed in the control plants (CK) at 168 h p.i. Differences were assessed using Student's t‐tests, and asterisks indicate P < 0.05.
Figure 8.
Transcript levels of some selected genes involved in cAMP‐PKA pathway of Pst after Pst infection. PsRAS1, GTPase Ras1; PsRAS2, GTPase Ras2; PsCPK2, catalytic subunits 2 of PKA; PsRPK, regulatory subunit of PKA; PsPrf1, HMG box transcription factor. Values represent the means ± standard error of three independent assays. Differences were assessed using Student's t‐tests, and asterisks indicate P < 0.05, and double asterisks indicate P < 0.01.
Discussion
To compensate for the limited amount of germplasm resources and rapid loss of resistance to fungal diseases, several new approaches have emerged in recent years. Many previous studies have experimentally validated that RNAi is a promising approach for durable control of pathogenic fungi (Chen et al., 2016; Cheng et al., 2015; Ghag et al., 2014; Nowara et al., 2010). In the rust fungi, the RNAi approach has been developed through BSMV (Yin et al., 2011). The Agrobacterium tumefaciens‐mediated transient RNAi assay has been developed in wheat to target P. triticina pathogenicity genes (Panwar et al., 2013a). But, complete resistance has rarely been achieved in wheat, and it is therefore significant that transgenic resistance to Pst was identified to the fourth generation in our study. To our knowledge, this is the first reported transgenic wheat with durable disease resistance against Pst.
A few studies have proved that expression of dsRNA targeted at fungal genes in wheat leads to increased resistance against phytopathogenic fungi (Chen et al., 2016; Cheng et al., 2015). Usually, true transgenic lines in hexaploid wheat are available until the T3 generation (Lee et al., 2012), and then, we generated T4 generation for further investigation. By analysing the T4 generation, we confirmed by northern blot analysis that transgene‐derived siRNAs accumulate in the resistant transgenic plants. Moreover, those plants had the strongest reduction in mRNA and corresponding decrease in susceptibility. Southern blot analysis indicated that each resistant transgenic line contains a single copy derived from independent transformations. Unlike the resistance of germplasm resources, the reduction in the disease phenotype does not depend on race‐specific resistance (Johnson, 1988) largely because of the high conservation of PsCPK1 in most isolates (data not shown), including the popular isolates CYR32 and V26 (Huang et al., 2014; Ren et al., 2015; Yin et al., 2009). Moreover, we found that transient silencing by BSMV‐HIGS is maintained for only 2 weeks, as indicated by Yin et al. (2011) and Miller et al. (2012), whereas the transgenic plants show a slow but continuous weakening of Pst silencing. Our results suggest that transgenic wheat has the potential to reduce the severity of stripe rust of wheat.
To further understand the mechanism after silencing the PsCPK1, we investigated the transcript levels of some important genes in the cAMP‐PKA pathway and the other subunit genes of the PKA complex. Ras proteins function at upstream of mitogen‐activated protein kinase (MAPK) or cAMP‐PKA pathway (Bluhm et al., 2007; Park et al., 2006). In Pst, PsRas2 is required for Pst pathogenicity, but not for PsRas1 (Cheng et al., 2016). PsRas2 and PsRas1 were significantly reduced after PsCPK1 silencing, perhaps through the feedback loop of the cAMP (Figure S7). The genes PsCPK2 and PsRPK, encoding another catalytic subunit and the regulatory subunit of the PKA complex, were down‐regulated at 168 h p.i. Interaction of the catalytic subunits of PKA with the regulatory subunit of PKA was activated by cAMP. Silencing of PsCPK1 may destroy the structure of the PKA complex. As Prf1 is required for cell fusion and filamentous growth (Hartmann et al., 1999) and the activity of Prf1 is controlled by PKA phosphorylation (Zarnack et al., 2008), a domino effect may appear in the transgenic plants after silencing of the PsCPK1. Together with these results, PsCPK1 plays important roles in the cAMP‐PKA pathway and silencing the PsCPK1 will disrupt the chain of cAMP‐PKA pathway after infection by Pst (Figure S7).
In the process of the application of RNAi methods in organisms, the off‐target effects maybe occur when the siRNA is partially complementary to one or more cellular mRNAs except the target (Jackson et al., 2003; de Souza, 2014). Despite the high conservation of CPK1 in different fungi, the PsCPK1 is sequence specific among its homologues in pathogenic fungi at the nucleotide level. Additionally, the homologous PsCPK1 gene in wheat shares up to 30% identity at the nucleotide sequence level within the coding regions. The possibility of off‐target effects was analysed by Si‐Fi software and no effective hits were found (Table S4). These off‐target effects can show no obvious difference in the growth of wheat (Figure S8). Furthermore, the off‐target effect could be more pronounced in rust fungi, which might have resulted in the disease suppression.
HIGS has been proved to be a novel tool to reveal gene function in obligate biotrophic fungus (Panwar et al., 2013a; Yin et al., 2011) and offers the potential of disease control (Nowara et al., 2010). In this study, wheat plants expressing the RNAi constructs showed strong and genetically stable resistance to Pst in the fourth generation. Our results indicated that PsCPK1 is an excellent target to generate durable genetic resistance against Pst and provides a potential reservoir of novel resistance resources of wheat against rust fungi. But the application of this material in agricultural production obviously requires more field trials. With the rapid development of various sequencing technologies, quantitative proteomics and RNA‐seq analysis may be used in this material to understand the mechanism of the cAMP‐mediated signal transduction pathway. The improved understanding will guide the development of genetic breeding, which could contribute to environmentally sustainable agriculture.
Experimental procedures
Biological materials, growth condition, fungal inoculation
The wheat cv. Suwon11 (Su11) and the Pst virulent isolate CYR32 were used in this study (Guo et al., 2011). Pst isolate CYR32 was maintained and propagated on susceptible wheat cv. Mingxian169. The germplasm used for gene transformation was XN1376, a high‐yielding and early‐maturing winter wheat variety. Plant cultivation and fungal inoculation were carried out following the procedures and conditions previously described (Kang et al., 2002).
Plasmid construction and plant transformation
To generate pMCG161‐CPK1, the PCR was performed to obtain the product with the primers PsCPK1‐F and PsCPK1‐R (Table S3). The amplified fragment was subsequently cloned into the AscI and AvrII and AsiSI and SpeI sites of plasmid pMCG161. Then, the CPK1+‐intro‐CPK1‐gene from T‐simple (TaKaRa, Tokyo, Japan) was cut with AscI and AvrII and ligated into the pMCG161 with AscI and AvrII.
Sequence alignments and polymorphism analysis
One cDNA clone encoding PsCPK1 (GenBank accession no. KY346510)was designed with special primer CPK1‐F/R (Table S3) according to the sequence from the cDNA library during wheat–Pst interaction (Ma et al., 2009). PsCPK1 sequence was firstly analysed using BLAST search and ORF Finder, and then, the conserved domain of PsCPK1 was detected with InterProScan and ScanProsite (Guo et al., 2013). Multiple sequence alignment was implemented with DNAMAN6.0 (Lynnon BioSoft, Quebec, Canada) and CLUSTALX2.0 (Chenna et al., 2003). The phylogenetic tree was constructed with the Mega 6.0 software (Tamura et al., 2013).
RNA extraction, cDNA synthesis and qRT‐PCR
To evaluate the transcript levels of PsCPK1 in response to Pst infection, wheat leaves were sampled at 0, 6, 12, 18, 24, 36, 48, 72, 120, 144, 216 and 264 h p.i. according to previous microscopic observations of wheat–Pst interaction (Wang et al., 2007). To determine the efficiency of BSMV‐mediated HIGS, the fourth leaves of PsCPK1‐knockdown plants were sampled at 24, 48 and 120 h p.i. with Pst. Total RNA was isolated with the Trizol reagent (Invitrogen, Carlsbad, CA), and the first‐strand cDNA was synthesized for qRT‐PCR. qRT‐PCR was performed with a 7500 Real‐Time PCR System (Applied Biosystems, Foster City, CA), and PCR conditions were used as previously described (Liu et al., 2012). The Pst translation elongation factor 1 (PsEF1) gene was used as the internal reference for normalization (Guo et al., 2011). The transcript levels of PsCPK1 and other genes in this study were assayed by the comparative 2−ΔΔCT method (Livak and Schmittgen, 2001).
Complementation of the M. oryzae cpkA mutant with PsCPK1
To perform complementation assays, the full length of the PsCPK1 gene was obtained with primers CPK1‐CM‐F and CPK1‐CM‐R (Table S3). Then, the PsCPK1 gene was cotransformed with the vector pFL2 into S. cerevisiae strain XK1‐25 to achieve plasmid pMoPsCPK1. The plasmid pMoPsCPK1 was transformed into protoplasts of the M. oryzae cpkA mutant DF51. To confirm the PsCPK1 gene integrated into the M. oryzae genome, we isolated the resultant transformants and verified them by PCR with primers CPK1‐CM‐F and CPK1‐CM‐R. The assays for appressorium formation and plant infection were performed as previously described (Guo et al., 2011).
BSMV‐mediated PsCPK1 gene silencing
To further determine the role of PsCPK1 during Pst infection, the PsCPK1 gene was silenced with the BSMV‐HIGS system. To make sure the specificity for PsCPK1 silencing, a 224‐bp fragment of PsCPK1 in the 5′UTR named PsCPK1‐1as and a 296‐bp fragment of PsCPK1 in the 3′UTR named PsCPK1‐2as were cloned with primers Higs‐CPK1‐1as‐F, Higs‐CPK1‐1as‐R, Higs‐CPK1‐2as‐F and Higs‐CPK1‐2as‐R (Table S3) and inserted into the virus plasmid. The BSMV RNAs were prepared in vitro from linearized plasmid γ‐TaPDSas, γ‐PsCPK1‐1as, γ‐PsCPK1‐2as, γ, α, β using the Message T7 in vitro transcription kit (Ambion, Austin, TX). The wheat leaves were used for inoculation with BSMV according to the procedures as previously described (Guo et al., 2013; Scofield et al., 2005). After inoculation with BSMV at the second leaf stage, wheat seedlings were maintained in a growth chamber at 23 ± 2 °C and examined for symptoms. In all experiments, the recombinant virus BSMV:TaPDSas was applied as a positive control. When the photobleaching phenotype was observed, the fourth leaves of PsCPK1 silencing group were inoculated with urediospores of Pst isolate CYR32. The resistant or susceptible phenotypes were visible at 15 dpi.
Histological observations of Pst growth and wheat response
The wheat leaves inoculated with BSMV at 24, 48 and 120 h p.i. and transgenic wheat leaves at 48 and 168 h p.i. were collected for histological observation as previously described (Guo et al., 2013). Stained leaf segments were fixed and cleared in ethanol/acetic acid (1:1 v/v). Autofluorescence of attacked mesophyll cells was observed as a necrotic death area with the Olympus BX‐51 microscope (Olympus, Tokyo, Japan). Infection sites and lengths of infection hyphae were measured under the blue light excitation. Fifty infection sites were examined on each randomly selected leaf segment per treatment for the measurement of fungal structures.
Northern blotting analysis
Northern blotting was used to detect the accumulation of small RNAs as described previously (Zheng et al., 2007). The total RNA from 12‐day‐old seedlings was extracted using Trizol reagent (Invitrogen). The poly(ethylene glycol) enrichment method was to obtain small RNA (Zheng et al., 2007). The random priming method was used to label the probe for the detection of mRNA transcripts. The primers NB‐CPK1‐F and NB‐CPK1‐R used to obtain the probe are listed in Table S3.
Southern blotting analysis
A CTAB‐based method was used to isolate total genomic DNA from 12‐day‐old seedlings (Hormaza, 2002). Aliquots (20 μg) of genomic DNA were digested overnight at 37 °C with BamHI and XhoI, fractionated in 0.8% (w/v) agarose gel and blotted onto a nylon membrane (Biotrace, Gelman Sciences, Ann Arbor, MI) according to the protocols previously described (Sambrook and Russell, 1989). The 302‐bp cDNA fragment was obtained by RT‐PCR with primer SB‐CPK1‐F/R (Table S3) and then used as the probe. The membrane was hybridized with the probe labelled with [α‐32P] dCTP. Hybridization, posthybridization washes and signal detection were carried out as described for northern blotting (Sambrook and Russell, 1989), but the hybridization temperature was adjusted to 65 °C.
Statistical analysis
Statistical testing was performed with the statistical software version package of IBM SPSS Statistics 21 (IBM SPSS Statistics, IBM Corporation, Armonk, NY). The data were tested by Student's t‐test (P < 0.05 or P < 0.01).
Supporting information
Figure S1 Amino acid sequence alignments of PsCPK1 with other fungal catalytic subunits of PKA.
Figure S2 Histological observation of fungal growth in PsCPK1‐knockdown wheat plants after inoculation with Pst isolate CYR32.
Figure S3 Bioassay of the transgenic wheat plants for Pst resistance.
Figure S4 The expression of the small RNA in T4 generation lines was analysed by RNA gel blotting.
Figure S5 Transcript profiles of PsCPK1 in the leaves of wheat cultivar XN1376 infected by Pst isolate CYR32.
Figure S6 Standard curves generated for the absolute quantification of Pst (A) and wheat (B).
Figure S7 Schematic presentation of possible HIGS mechanisms involved in PKA pathway.
Figure S8 No significant difference was observed for the growth between transgenic and control wheat lines.
Table S1 Transformation pipeline and efficiency for producing transgenic lines used in this study.
Table S2 Disease severity scale of wheat stripe rust.
Table S3 Primers designed for PsCPK1 research.
Table S4 Prediction off‐target transcripts of PsCPK1 gene.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31620103913 and 31371889) and the National Basic Research Program of China (2013CB127700). The authors declare no conflict of interest.
Contributor Information
Zhensheng Kang, Email: kangzs@nwsuaf.edu.cn.
Jun Guo, Email: guojunwgq@nwafu.edu.cn.
References
- Bahn, Y. and Sundstrom, P. (2001) CAP1, an adenylate cyclase‐associated protein gene, regulates bud‐hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans . J. Bacteriol. 183, 3211–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm, B.H. , Zhao, X. , Flaherty, J.E. , Xu, J. and Dunkle, L.D. (2007) RAS2 regulates growth and pathogenesis in Fusarium graminearum . Mol. Plant Microbe Interact. 20, 627–636. [DOI] [PubMed] [Google Scholar]
- Castel, S.E. and Martienssen, R.A. (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Wellings, C. , Chen, X. , Kang, Z. and Liu, T. (2013) Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici . Mol. Plant Pathol. 15, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Kastner, C. , Nowara, D. , Oliveira‐Garcia, E. , Rutten, T. , Zhao, Y. , Deising, H.B. et al (2016) Host‐induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 67, 4979–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W. , Song, X. , Li, H. , Cao, L. , Sun, K. , Qiu, X. , Xu, Y. et al (2015) Host‐induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Wang, W. , Yao, J. , Huang, L. , Voegele, R.T. , Wang, X.J. and Kang, Z.S. (2016) Two distinct Ras genes from Puccinia striiformis exhibit differential roles in rust pathogenicity and cell death. Environ. Microbiol. 18, 3910–3922. [DOI] [PubMed] [Google Scholar]
- Chenna, R. , Sugawara, H. , Koike, T. , Lopez, R. , Gibson, T.J. , Higgins, D.G. and Thompson, J.D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, C.A. and Heitman, J. (2001) Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25, 349–364. [DOI] [PubMed] [Google Scholar]
- D'Souza, C.A. , Alspaugh, J.A. , Yue, C. , Harashima, T. , Cox, G.M. , Perfect, J.R. and Heitman, J. (2001) Cyclic AMP‐dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans . Mol. Cell. Biol. 21, 3179–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis, S. , Cuomo, C.A. , Lin, Y. , Aerts, A. , Tisserant, E. , Veneault‐Fourrey, C. , Joly, D.L. et al (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl Acad. Sci. USA, 108, 9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrenberger, F. , Wong, K. and Kronstad, J.W. (1998) Identification of a cAMP‐dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis . Proc. Natl Acad. Sci. USA, 95, 5684–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A. , Xu, S. , Montgomery, M.K. , Kostas, S.A. , Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. and Gurr, S.J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature, 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, K.K. and Rhodes, J.C. (2012) Protein kinase A and fungal virulence. Virulence, 3, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghag, S.B. , Shekhawat, U.K.S. and Ganapathi, T.R. (2014) Host‐induced post‐transcriptional hairpin RNA‐mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 12, 541–553. [DOI] [PubMed] [Google Scholar]
- Govindarajulu, M. , Epstein, L. , Wroblewski, T. and Michelmore, R.W. (2015) Host‐induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883. [DOI] [PubMed] [Google Scholar]
- Guo, J. , Dai, X. , Xu, J. , Wang, Y. , Bai, P. , Liu, F. , Duan, Y. et al (2011) Molecular characterization of a Fus3/Kss1 type MAPK from Puccinia striiformis f. sp. tritici, PsMAPK1. PLoS ONE, 6, e21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J. , Bai, P. , Yang, Q. , Liu, F. , Wang, X. , Huang, L. and Kang, Z. (2013) Wheat zinc finger protein TaLSD1, a negative regulator of programmed cell death, is involved in wheat resistance against stripe rust fungus. Plant Physiol. Biochem. 71, 164–172. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Hartmann, H.A. , Krüger, J. , Lottspeich, F. and Kahmann, R. (1999) Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell, 11, 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassebrauk, K. and Schroeder, J. (1964) Studies on the germination of yellow rust urediospores In Proceedings of the First European and Mediterranean Cereal Rusts Conference (Macer R. C. F. and Wolfe M. S., eds.), pp. 12–18. Cambridge UK: Plant Breeding Institute. [Google Scholar]
- Hellens, R.P. , Allan, A.C. , Friel, E.N. , Bolitho, K. , Grafton, K. , Templeton, M.D. , Karunairetnam, S. et al (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaza, J.I. (2002) Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theor. Appl. Genet. 104, 321–328. [DOI] [PubMed] [Google Scholar]
- Hu, S. , Zhou, X. , Gu, X. , Cao, S. , Wang, C. and Xu, J. (2014) The cAMP‐PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum . Mol. Plant Microbe Interact. 27, 557–566. [DOI] [PubMed] [Google Scholar]
- Huang, Q. , Li, X. , Chen, W.Q. , Xiang, Z.P. , Zhong, S.F. , Chang, Z.J. , Zhang, M. et al (2014) Genetic mapping of a putative Thinopyrum intermedium‐derived stripe rust resistance gene on wheat chromosome 1B. Theor. Appl. Genet. 127, 843–853. [DOI] [PubMed] [Google Scholar]
- Huvenne, H. and Smagghe, G. (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L. , Bartz, S.R. , Schelter, J. , Kobayashi, S.V. , Burchard, J. , Mao, M. , Li, B. et al (2003) Expression profiling reveals off‐target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637. [DOI] [PubMed] [Google Scholar]
- Johnson, R. (1988) Durable resistance to yellow (stripe) rust in wheat and its implications in plant breeding In Breeding Strategies for Resistance to the Rusts of Wheat (Simmonds N. W. and Rajaram S., eds.), pp. 63–75. Mexico D.F.: CIMMYT. [Google Scholar]
- Kang, Z. , Huang, L. and Buchenauer, H. (2002) Ultrastructural changes and localization of lignin and callose in compatible and incompatible interactions between wheat and Puccinia striiformis . J. Plant Dis. Prot. 109, 25–37. [Google Scholar]
- Kang, Z.S. , Wang, Y. , Huang, L.L. , Wei, G.R. and Zhao, J. (2003) Histology and ultrastructure of incompatible combination between Puccinia striiformis and wheat cultivars with resistance of low reaction type. Sci. Agric. Sin. 36, 1026–1031. [Google Scholar]
- Lee, W. , Hammond‐Kosack, K.E. and Kanyuka, K. (2012) Barley stripe mosaic virus‐mediated tools for investigating gene function in cereal plants and their pathogens: virus‐induced gene silencing, host‐mediated gene silencing, and virus‐mediated overexpression of heterologous protein. Plant Physiol. 160, 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Line, R.F. and Qayoum, A. (1992) Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America, pp. 1968–1987. Technical Bulletin (USA).
- Liu, F. , Guo, J. , Bai, P. , Duan, Y. , Wang, X. , Cheng, Y. , Feng, H. et al (2012) Wheat TaRab7 GTPase is part of the signaling pathway in responses to stripe rust and abiotic stimuli. PLoS ONE, 7, e37146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Huang, X. , Wang, X. , Chen, X. , Qu, Z. , Huang, L. and Kang, Z. (2009) Identification of expressed genes during compatible interaction between stripe rust (Puccinia striiformis) and wheat using a cDNA library. BMC Genom. 10, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.C. , Miyata, K. , Brown, S.J. and Tomoyasu, Y. (2012) Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS ONE, 7, e47431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, T.K. and Dean, R.A. (1995) The cAMP‐dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea . Plant Cell, 7, 1869–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. , Hensel, G. et al (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. and Heitman, J. (1999) Cyclic AMP‐dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae . Mol. Cell. Biol. 19, 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar, V. , McCallum, B. and Bakkeren, G. (2013a) Endogenous silencing of Puccinia triticina pathogenicity genes through in planta‐expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 73, 521–532. [DOI] [PubMed] [Google Scholar]
- Panwar, V. , McCallum, B. and Bakkeren, G. (2013b) Host‐induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 81, 595–608. [DOI] [PubMed] [Google Scholar]
- Park, G. , Xue, C. , Zhao, X. , Kim, Y. , Orbach, M. and Xu, J. (2006) Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea . Plant Cell, 18, 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y. , Li, S. , Xia, X. , Zhou, Q. , He, Y. , Wei, Y. , Zheng, Y. et al (2015) Molecular mapping of a recessive stripe rust resistance gene yrMY37 in Chinese wheat cultivar Mianmai 37. Mol. Breeding, 35, 1–9. [Google Scholar]
- Robertson, L.S. and Fink, G.R. (1998) The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl Acad. Sci. USA, 95, 13783–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (1989) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scofield, S.R. , Huang, L. , Brandt, A.S. and Gill, B.S. (2005) Development of a virus‐induced gene‐silencing system for hexaploid wheat and its use in functional analysis of the Lr21‐mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza, N. (2014) Genetics: more specific CRISPR editing. Nat. Methods, 11, 712. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein, J.M. and De Winde, J.H. (1999) Novel sensing mechanisms and targets for the cAMP‐protein kinase A pathway in the yeast Saccharomyces cerevisiae . Mol. Microbiol. 33, 904–918. [DOI] [PubMed] [Google Scholar]
- Toda, T. , Cameron, S. , Sass, P. , Zoller, M. and Wigler, M. (1987) Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP‐dependent protein kinase. Cell, 50, 277–287. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. and Fagard, M. (2001) Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17, 29–35. [DOI] [PubMed] [Google Scholar]
- Voegele, R.T. and Mendgen, K. (2003) Rust haustoria: nutrient uptake and beyond. New Phytol. 159, 93–100. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Huang, L. , Buchenauer, H. , Han, Q. , Zhang, H. and Kang, Z. (2007) Histochemical studies on the accumulation of reactive oxygen species (O2− and H2O2) in the incompatible and compatible interaction of wheat‐Puccinia striiformis f. sp. tritici . Physiol. Mol. Plant Pathol. 71, 230–239. [Google Scholar]
- Xu, J. , Urban, M. , Sweigard, J.A. and Hamer, J.E. (1997) The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol. Plant Microbe Interact. 10, 187–194. [Google Scholar]
- Xu, J. , Linning, R. , Fellers, J. , Dickinson, M. , Zhu, W. , Antonov, I. , Joly, D.L. et al (2011) Gene discovery in EST sequences from the wheat leaf rust fungus Puccinia triticina sexual spores, asexual spores and haustoria, compared to other rust and corn smut fungi. BMC Genom. 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, C. , Chen, X. , Wang, X. , Han, Q. , Kang, Z. and Hulbert, S.H. (2009) Generation and analysis of expression sequence tags from haustoria of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici . BMC Genom. 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, C. , Jurgenson, J.E. and Hulbert, S.H. (2011) Development of a host‐induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici . Mol. Plant Microbe Interact. 24, 554–561. [DOI] [PubMed] [Google Scholar]
- Zarnack, K. , Eichhorn, H. , Kahmann, R. and Feldbrügge, M. (2008) Pheromone‐regulated target genes respond differentially to MAPK phosphorylation of transcription factor Prf1. Mol. Microbiol. 69, 1041–1053. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Zhu, J. , Kapoor, A. and Zhu, J.K. (2007) Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26, 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, W.M. , Huang, L.L. , Huang, J.Q. , Wang, X.J. , Chen, X.M. , Zhao, J. , Guo, J. et al (2013) High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 4, 2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Amino acid sequence alignments of PsCPK1 with other fungal catalytic subunits of PKA.
Figure S2 Histological observation of fungal growth in PsCPK1‐knockdown wheat plants after inoculation with Pst isolate CYR32.
Figure S3 Bioassay of the transgenic wheat plants for Pst resistance.
Figure S4 The expression of the small RNA in T4 generation lines was analysed by RNA gel blotting.
Figure S5 Transcript profiles of PsCPK1 in the leaves of wheat cultivar XN1376 infected by Pst isolate CYR32.
Figure S6 Standard curves generated for the absolute quantification of Pst (A) and wheat (B).
Figure S7 Schematic presentation of possible HIGS mechanisms involved in PKA pathway.
Figure S8 No significant difference was observed for the growth between transgenic and control wheat lines.
Table S1 Transformation pipeline and efficiency for producing transgenic lines used in this study.
Table S2 Disease severity scale of wheat stripe rust.
Table S3 Primers designed for PsCPK1 research.
Table S4 Prediction off‐target transcripts of PsCPK1 gene.


