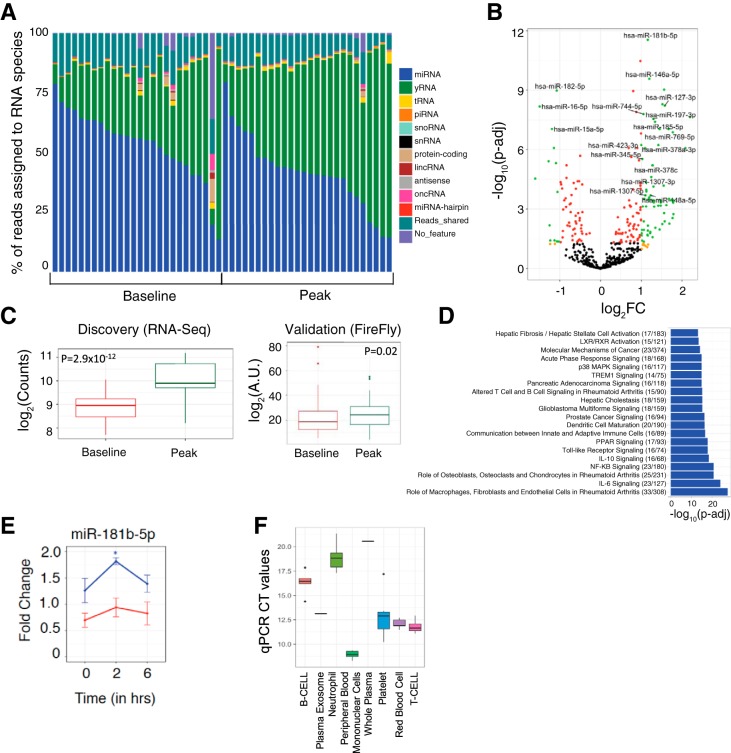
Fig. 1.
A: stacked-bar plot representing the percentage of reads assigned to various RNA species. B: volcano plot of the differentially expressed miRNAs. Green dots represent miRNAs that are differentially expressed at an absolute value (fold change) of ≥2 and false discovery rate adjusted <0.05 (Benjamini-Hochberg). C: plasma expression of miR-181b-5p in discovery and validation cohorts (P < 0.05 for validation using a two-sided Wilcoxon rank-sum test). D: top 20 canonical biological pathways from Ingenuity Pathway Analysis specified by miRNAs dysregulated during acute maximal exercise. The numbers in parentheses refer to the number of genes targeted by differentially expressed miRNAs over the total number of genes in a given pathway. The P values obtained from a two-tailed Fisher exact test were adjusted for multiple testing (Benjamini-Hochberg). E: miR-181b-5p fold change in murine skeletal muscle tissue after acute exercise in old (red; n = 3) and young (blue; n = 3) mice with respect to age-matched control (n = 3) mice as a function of time (0: before exercise; 2 and 6 h: after exercise). A two-tailed t-test was used to determine significance between the experimental group (exercised mouse) and age-matched control mice. *Bonferroni-adjusted P value < 0.05. F: miR-181b-5p expression in constituent cells in blood separated by flow sorting. Targeted quantitative PCR (qPCR) using the Qiagen miScript platform was performed, and the computed tomography (CT) values are presented as a box plot for 6 replicates of each cell type.